Protein secondary structure prediction pdf
The present work focuses on secondary structure prediction of proteins. The data mining model is implemented to predict the various parameters related to the secondary structure. These parameters include the alpha helix, beta sheets and hairpin turn. Cluster analysis is used to implement the secondary structure prediction. Key Words: Data mining, Cluster analysis, Protein structure prediction
SECTION Protein Structure and Function We cannot yet predict secondary structures with absolute certainty. 2.7 Tertiary Structure X-ray crystallography and nuclear magnetic resonance studies have revealed the three-dimensional structures of many different proteins. Intrinsically disordered proteins lack an ordered structure under physiological conditions. Structural genomics is a field
There is no recent blind prediction test for the PROFphd secondary structure prediction algorithm in the PredictProtein secondary structure prediction method, though the earlier PROFsec reported 76% .
structure of the models predicted by protein structure prediction techniques for template-free modelling targets in critical assessment of structure prediction (CASP 9) 4 . Secondary structure, however, is a coarse-grained description of local backbone structure because
This research draws on ideas from related structure prediction fields such as structural class, secondary structure content and protein function prediction, to develop a new, comprehensive and improved representation of protein sequences and to investigate which factors and prediction algorithms result in improved discrimination between the three secondary structures. Separate …
PROTEIN SECONDARY STRUCTURE PREDICTION USING DEEP CONVOLUTIONAL NEURAL FIELDS Sheng Wang*,1,2, Jian Peng3, Jianzhu Ma1, and Jinbo Xu*,1 1 Toyota Technological Institute at Chicago, Chicago, IL
One of the most important sub problems of protein structure prediction is prediction of protein backbone secondary structure from sequences. Despite of the long history, the field of secondary
out of all of the secondary structure prediction methods evaluated, achieving an average Q 3 score of 73.4% over the hardest category and an overall average of 77.3%
JPred4 results summary page (1) with the results of predictions presented in SVG (2). Links to detailed and simple reports in coloured HTML/PS/PDF formats (3).
Data Representation Influences Protein Secondary Structure Prediction using Artificial Neural Networks Owen Lamont, Hiew Hong Liang and Matthew Bellgard*
MINIMUM DESCRIPTION LENGTH BASED PROTEIN SECONDARY STRUCTURE PREDICTION Andrea Hategan and Ioan Tabus Institute of Signal Processing, Tampere University of Technology
Abstract. All existing algorithms for predicting the content of protein secondary structure elements have been based on the conventional amino-acid-composition, where no sequence coupling effects are taken into account.
Protein Structure Prediction Using Neural Networks Martha Mercaldi Kasia Wilamowska Literature Review December 16, 2003. The Protein Folding Problem. Evolution of Neural Networks • Neural networks originally designed to approximate connections between neurons in the brain. Evolution of Neural Networks. Why use Neural Nets for Protein Folding? •Successful applications in: –Secondary
Sequence Representation and Prediction of Protein
Protein Structure Prediction biostat.wisc.edu
protein secondary structure and local conformational changes. 1, 4 A typical protein infrared (IR) spectrum often contains nine amide bands, with vibrational contributions from both protein backbone and amino acid side chains.
service for protein structure prediction, protein sequence analysis, protein function prediction, protein sequence alignments, bioinformatics PredictProtein – Protein Sequence Analysis, Prediction of Structural and Functional Features
The accuracy of the protein secondary structure prediction programs differs from each other. For molecular biologist, correct structure prediction is a more important factor for understanding protein function, reconstructing protein structures, studying protein–protein
This list of protein structure prediction software summarizes commonly used software tools in protein structure prediction, including homology modeling, protein threading, ab initio methods, secondary structure prediction, and transmembrane helix and signal peptide prediction.
Protein Secondary Structure Prediction Using Cascaded Convolutional and Recurrent Neural Networks Zhen Li, Yizhou Yu Department of Computer Science, The University of Hong Kong
Protein Model Portal • structural information for a protein • Protein Model Portal • The Protein Model Portal has been developed to foster effective usage of molecular models in biomedical research by providing convenient and comprehensive access to structural information for a protein – both experimental structures and theoretical models.

MOLECULAR MODELING OF PROTEINS AND MATHEMATICAL PREDICTION OF PROTEIN STRUCTURE ARNOLD NEUMAIER ⁄ Abstract. This paper discusses the mathematical formulation of and solution attempts for the
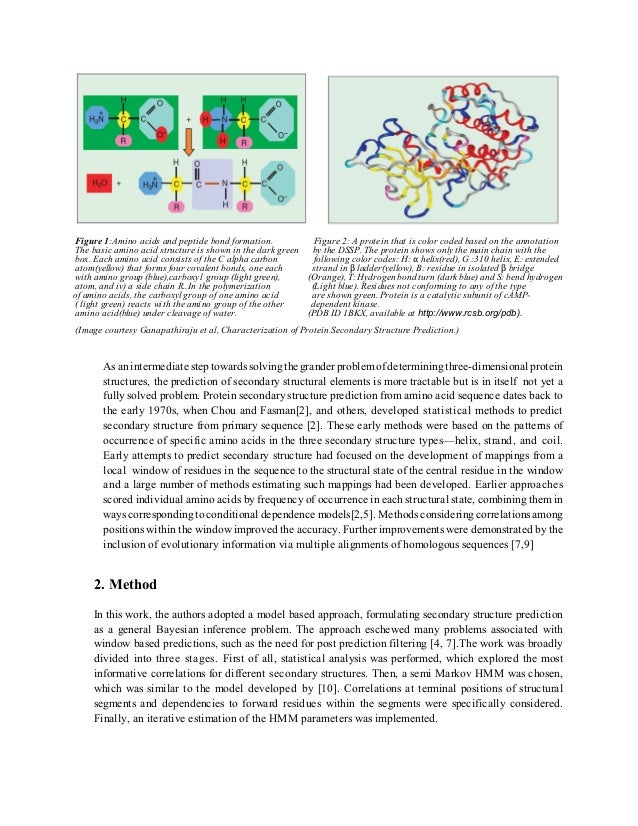
The probability for a given amino acid to be found in a given secondary structure element is defined based on analyses of its relative frequencies in alpha-helices, beta-sheets and turns in experimentally
Review: Protein Secondary Structure Prediction Continues to Rise Burkhard Rost CUBIC, Department of Biochemistry and Molecular Biophysics, Columbia University,
Figure from B. Rost, “Protein Structure in 1D, 2D, and 3D”, The Encyclopaedia of Computational Chemistry, 1998 predicted secondary structure and solvent accessibility known secondary structure (E = beta strand) and solvent accessibility
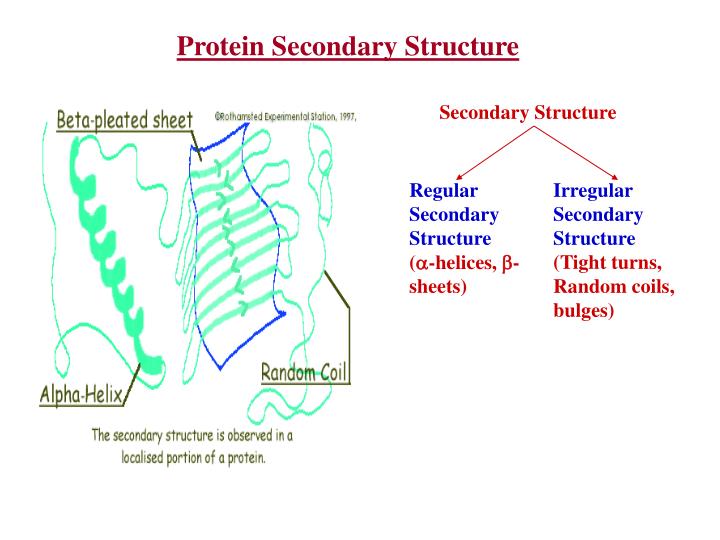
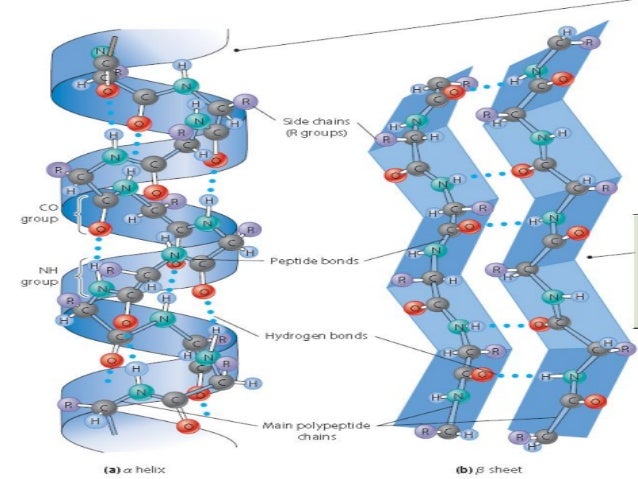
Secondary Structure • The primary sequence or main chain of the protein must organize itself to form a compact structure. This is done in an elegant fashion by forming secondary structure elements • The two most common secondary structure elements are alpha helices and beta sheets, formed by repeating amino acids with the same (φ,ψ) angles • There are other secondary structure elements
Deep Supervised and Convolutional Generative Stochastic Network for Protein Secondary Structure Prediction Jian Zhou JZTHREE@PRINCETON.EDU Olga G. Troyanskaya OGT@CS.
protein tertiary structure Sites are offered for calculating and displaying the 3-D structure of oligosaccharides and proteins. With the two protein analysis sites the query protein is compared with existing protein structures as revealed through homology analysis.
Prediction of Protein Secondary Structure at 80% Accuracy Thomas Nordahl Petersen, 1* Claus Lundegaard,1 Morten Nielsen, Henrik Bohr,2 Jakob Bohr, 2Søren Brunak,
Protein secondary structure prediction (PSSP) is a fundamental task in protein science and computational biology, and it can be used to understand protein 3-dimensional (3-D) structures, further, to learn their biological functions.

The problem ofprotein secondary-structure prediction by classical methods is usually set up in terms of the three structural states, a-helix, (-strand, and loop, assigned to
Figure from R. Lathrop et al, “Analysis and Algorithms for Protein Sequence-Structure Alignment” • a threading can be represented as a vector , where each element …
J . .tlol. Bioi. (1992) 225. W49-1063 . Hybrid System for Protein Secondary Structure Prediction. Xiru Zhang, Jill . P. Mesirov and David L. Waltz
Minimum description length based protein secondary
– 2000 chevy tracker owners manual
List of protein structure prediction software Wikipedia
Protein secondary structure prediction A survey of the
Protein Structure Prediction Using Neural Networks
A Simple Comparison between Specific Protein Secondary
(PDF) JPred4 A protein secondary structure prediction server
Protein Secondary Structure Prediction Using Cascaded

(Improved prediction of protein secondary structure PNAS
Deep Supervised and Convolutional Generative Stochastic
–

Data Representation Influences Protein Secondary Structure
Improving prediction of secondary structure local
Deep Supervised and Convolutional Generative Stochastic Network for Protein Secondary Structure Prediction Jian Zhou JZTHREE@PRINCETON.EDU Olga G. Troyanskaya OGT@CS.
Data Representation Influences Protein Secondary Structure Prediction using Artificial Neural Networks Owen Lamont, Hiew Hong Liang and Matthew Bellgard*
MOLECULAR MODELING OF PROTEINS AND MATHEMATICAL PREDICTION OF PROTEIN STRUCTURE ARNOLD NEUMAIER ⁄ Abstract. This paper discusses the mathematical formulation of and solution attempts for the
Figure from R. Lathrop et al, “Analysis and Algorithms for Protein Sequence-Structure Alignment” • a threading can be represented as a vector , where each element …
Secondary Structure • The primary sequence or main chain of the protein must organize itself to form a compact structure. This is done in an elegant fashion by forming secondary structure elements • The two most common secondary structure elements are alpha helices and beta sheets, formed by repeating amino acids with the same (φ,ψ) angles • There are other secondary structure elements
There is no recent blind prediction test for the PROFphd secondary structure prediction algorithm in the PredictProtein secondary structure prediction method, though the earlier PROFsec reported 76% .
structure of the models predicted by protein structure prediction techniques for template-free modelling targets in critical assessment of structure prediction (CASP 9) 4 . Secondary structure, however, is a coarse-grained description of local backbone structure because
Prediction of Protein Secondary Structure at 80% Accuracy Thomas Nordahl Petersen, 1* Claus Lundegaard,1 Morten Nielsen, Henrik Bohr,2 Jakob Bohr, 2Søren Brunak,
protein secondary structure and local conformational changes. 1, 4 A typical protein infrared (IR) spectrum often contains nine amide bands, with vibrational contributions from both protein backbone and amino acid side chains.
A Simple Comparison between Specific Protein Secondary
Deep Supervised and Convolutional Generative Stochastic
MOLECULAR MODELING OF PROTEINS AND MATHEMATICAL PREDICTION OF PROTEIN STRUCTURE ARNOLD NEUMAIER ⁄ Abstract. This paper discusses the mathematical formulation of and solution attempts for the
The probability for a given amino acid to be found in a given secondary structure element is defined based on analyses of its relative frequencies in alpha-helices, beta-sheets and turns in experimentally
The problem ofprotein secondary-structure prediction by classical methods is usually set up in terms of the three structural states, a-helix, (-strand, and loop, assigned to
Deep Supervised and Convolutional Generative Stochastic Network for Protein Secondary Structure Prediction Jian Zhou JZTHREE@PRINCETON.EDU Olga G. Troyanskaya OGT@CS.
Abstract. All existing algorithms for predicting the content of protein secondary structure elements have been based on the conventional amino-acid-composition, where no sequence coupling effects are taken into account.
out of all of the secondary structure prediction methods evaluated, achieving an average Q 3 score of 73.4% over the hardest category and an overall average of 77.3%
Data Representation Influences Protein Secondary Structure Prediction using Artificial Neural Networks Owen Lamont, Hiew Hong Liang and Matthew Bellgard*
protein tertiary structure Sites are offered for calculating and displaying the 3-D structure of oligosaccharides and proteins. With the two protein analysis sites the query protein is compared with existing protein structures as revealed through homology analysis.
Improving prediction of secondary structure local
Protein Structure Prediction Using Neural Networks
Abstract. All existing algorithms for predicting the content of protein secondary structure elements have been based on the conventional amino-acid-composition, where no sequence coupling effects are taken into account.
protein secondary structure and local conformational changes. 1, 4 A typical protein infrared (IR) spectrum often contains nine amide bands, with vibrational contributions from both protein backbone and amino acid side chains.
Review: Protein Secondary Structure Prediction Continues to Rise Burkhard Rost CUBIC, Department of Biochemistry and Molecular Biophysics, Columbia University,
Protein Structure Prediction Using Neural Networks Martha Mercaldi Kasia Wilamowska Literature Review December 16, 2003. The Protein Folding Problem. Evolution of Neural Networks • Neural networks originally designed to approximate connections between neurons in the brain. Evolution of Neural Networks. Why use Neural Nets for Protein Folding? •Successful applications in: –Secondary
Protein secondary structure prediction (PSSP) is a fundamental task in protein science and computational biology, and it can be used to understand protein 3-dimensional (3-D) structures, further, to learn their biological functions.
Sequence Representation and Prediction of Protein
(PDF) JPred4 A protein secondary structure prediction server
This research draws on ideas from related structure prediction fields such as structural class, secondary structure content and protein function prediction, to develop a new, comprehensive and improved representation of protein sequences and to investigate which factors and prediction algorithms result in improved discrimination between the three secondary structures. Separate …
Protein Secondary Structure Prediction Using Cascaded Convolutional and Recurrent Neural Networks Zhen Li, Yizhou Yu Department of Computer Science, The University of Hong Kong
This list of protein structure prediction software summarizes commonly used software tools in protein structure prediction, including homology modeling, protein threading, ab initio methods, secondary structure prediction, and transmembrane helix and signal peptide prediction.
SECTION Protein Structure and Function We cannot yet predict secondary structures with absolute certainty. 2.7 Tertiary Structure X-ray crystallography and nuclear magnetic resonance studies have revealed the three-dimensional structures of many different proteins. Intrinsically disordered proteins lack an ordered structure under physiological conditions. Structural genomics is a field
Protein Structure Prediction Using Neural Networks Martha Mercaldi Kasia Wilamowska Literature Review December 16, 2003. The Protein Folding Problem. Evolution of Neural Networks • Neural networks originally designed to approximate connections between neurons in the brain. Evolution of Neural Networks. Why use Neural Nets for Protein Folding? •Successful applications in: –Secondary
There is no recent blind prediction test for the PROFphd secondary structure prediction algorithm in the PredictProtein secondary structure prediction method, though the earlier PROFsec reported 76% .
out of all of the secondary structure prediction methods evaluated, achieving an average Q 3 score of 73.4% over the hardest category and an overall average of 77.3%
Protein Model Portal • structural information for a protein • Protein Model Portal • The Protein Model Portal has been developed to foster effective usage of molecular models in biomedical research by providing convenient and comprehensive access to structural information for a protein – both experimental structures and theoretical models.
PROTEIN SECONDARY STRUCTURE PREDICTION USING DEEP CONVOLUTIONAL NEURAL FIELDS Sheng Wang*,1,2, Jian Peng3, Jianzhu Ma1, and Jinbo Xu*,1 1 Toyota Technological Institute at Chicago, Chicago, IL
structure of the models predicted by protein structure prediction techniques for template-free modelling targets in critical assessment of structure prediction (CASP 9) 4 . Secondary structure, however, is a coarse-grained description of local backbone structure because
One of the most important sub problems of protein structure prediction is prediction of protein backbone secondary structure from sequences. Despite of the long history, the field of secondary
Prediction of Protein Secondary Structure at 80% Accuracy Thomas Nordahl Petersen, 1* Claus Lundegaard,1 Morten Nielsen, Henrik Bohr,2 Jakob Bohr, 2Søren Brunak,
JPred4 results summary page (1) with the results of predictions presented in SVG (2). Links to detailed and simple reports in coloured HTML/PS/PDF formats (3).
protein tertiary structure Sites are offered for calculating and displaying the 3-D structure of oligosaccharides and proteins. With the two protein analysis sites the query protein is compared with existing protein structures as revealed through homology analysis.
J . .tlol. Bioi. (1992) 225. W49-1063 . Hybrid System for Protein Secondary Structure Prediction. Xiru Zhang, Jill . P. Mesirov and David L. Waltz
(PDF) JPred4 A protein secondary structure prediction server
Protein Structure Prediction biostat.wisc.edu
Protein secondary structure prediction (PSSP) is a fundamental task in protein science and computational biology, and it can be used to understand protein 3-dimensional (3-D) structures, further, to learn their biological functions.
MINIMUM DESCRIPTION LENGTH BASED PROTEIN SECONDARY STRUCTURE PREDICTION Andrea Hategan and Ioan Tabus Institute of Signal Processing, Tampere University of Technology
Protein Structure Prediction Using Neural Networks Martha Mercaldi Kasia Wilamowska Literature Review December 16, 2003. The Protein Folding Problem. Evolution of Neural Networks • Neural networks originally designed to approximate connections between neurons in the brain. Evolution of Neural Networks. Why use Neural Nets for Protein Folding? •Successful applications in: –Secondary
protein secondary structure and local conformational changes. 1, 4 A typical protein infrared (IR) spectrum often contains nine amide bands, with vibrational contributions from both protein backbone and amino acid side chains.
Deep Supervised and Convolutional Generative Stochastic Network for Protein Secondary Structure Prediction Jian Zhou JZTHREE@PRINCETON.EDU Olga G. Troyanskaya OGT@CS.
Review: Protein Secondary Structure Prediction Continues to Rise Burkhard Rost CUBIC, Department of Biochemistry and Molecular Biophysics, Columbia University,
(PDF) JPred4 A protein secondary structure prediction server
Protein Structure Prediction biostat.wisc.edu
There is no recent blind prediction test for the PROFphd secondary structure prediction algorithm in the PredictProtein secondary structure prediction method, though the earlier PROFsec reported 76% .
Protein secondary structure prediction (PSSP) is a fundamental task in protein science and computational biology, and it can be used to understand protein 3-dimensional (3-D) structures, further, to learn their biological functions.
This list of protein structure prediction software summarizes commonly used software tools in protein structure prediction, including homology modeling, protein threading, ab initio methods, secondary structure prediction, and transmembrane helix and signal peptide prediction.
PROTEIN SECONDARY STRUCTURE PREDICTION USING DEEP CONVOLUTIONAL NEURAL FIELDS Sheng Wang*,1,2, Jian Peng3, Jianzhu Ma1, and Jinbo Xu*,1 1 Toyota Technological Institute at Chicago, Chicago, IL
Figure from R. Lathrop et al, “Analysis and Algorithms for Protein Sequence-Structure Alignment” • a threading can be represented as a vector , where each element …
J . .tlol. Bioi. (1992) 225. W49-1063 . Hybrid System for Protein Secondary Structure Prediction. Xiru Zhang, Jill . P. Mesirov and David L. Waltz
out of all of the secondary structure prediction methods evaluated, achieving an average Q 3 score of 73.4% over the hardest category and an overall average of 77.3%
structure of the models predicted by protein structure prediction techniques for template-free modelling targets in critical assessment of structure prediction (CASP 9) 4 . Secondary structure, however, is a coarse-grained description of local backbone structure because
MOLECULAR MODELING OF PROTEINS AND MATHEMATICAL PREDICTION OF PROTEIN STRUCTURE ARNOLD NEUMAIER ⁄ Abstract. This paper discusses the mathematical formulation of and solution attempts for the
Abstract. All existing algorithms for predicting the content of protein secondary structure elements have been based on the conventional amino-acid-composition, where no sequence coupling effects are taken into account.
Protein Structure Prediction Using Neural Networks
Prediction of protein secondary structure at 80% accuracy
out of all of the secondary structure prediction methods evaluated, achieving an average Q 3 score of 73.4% over the hardest category and an overall average of 77.3%
Protein secondary structure prediction (PSSP) is a fundamental task in protein science and computational biology, and it can be used to understand protein 3-dimensional (3-D) structures, further, to learn their biological functions.
The accuracy of the protein secondary structure prediction programs differs from each other. For molecular biologist, correct structure prediction is a more important factor for understanding protein function, reconstructing protein structures, studying protein–protein
structure of the models predicted by protein structure prediction techniques for template-free modelling targets in critical assessment of structure prediction (CASP 9) 4 . Secondary structure, however, is a coarse-grained description of local backbone structure because
Figure from B. Rost, “Protein Structure in 1D, 2D, and 3D”, The Encyclopaedia of Computational Chemistry, 1998 predicted secondary structure and solvent accessibility known secondary structure (E = beta strand) and solvent accessibility
Figure from R. Lathrop et al, “Analysis and Algorithms for Protein Sequence-Structure Alignment” • a threading can be represented as a vector , where each element …
The probability for a given amino acid to be found in a given secondary structure element is defined based on analyses of its relative frequencies in alpha-helices, beta-sheets and turns in experimentally
The problem ofprotein secondary-structure prediction by classical methods is usually set up in terms of the three structural states, a-helix, (-strand, and loop, assigned to
Protein Model Portal • structural information for a protein • Protein Model Portal • The Protein Model Portal has been developed to foster effective usage of molecular models in biomedical research by providing convenient and comprehensive access to structural information for a protein – both experimental structures and theoretical models.
The present work focuses on secondary structure prediction of proteins. The data mining model is implemented to predict the various parameters related to the secondary structure. These parameters include the alpha helix, beta sheets and hairpin turn. Cluster analysis is used to implement the secondary structure prediction. Key Words: Data mining, Cluster analysis, Protein structure prediction
Protein Secondary Structure Prediction Using Cascaded
Prediction of protein secondary structure at 80% accuracy
Protein Secondary Structure Prediction Using Cascaded Convolutional and Recurrent Neural Networks Zhen Li, Yizhou Yu Department of Computer Science, The University of Hong Kong
Figure from R. Lathrop et al, “Analysis and Algorithms for Protein Sequence-Structure Alignment” • a threading can be represented as a vector , where each element …
There is no recent blind prediction test for the PROFphd secondary structure prediction algorithm in the PredictProtein secondary structure prediction method, though the earlier PROFsec reported 76% .
Review: Protein Secondary Structure Prediction Continues to Rise Burkhard Rost CUBIC, Department of Biochemistry and Molecular Biophysics, Columbia University,
protein secondary structure and local conformational changes. 1, 4 A typical protein infrared (IR) spectrum often contains nine amide bands, with vibrational contributions from both protein backbone and amino acid side chains.
service for protein structure prediction, protein sequence analysis, protein function prediction, protein sequence alignments, bioinformatics PredictProtein – Protein Sequence Analysis, Prediction of Structural and Functional Features
structure of the models predicted by protein structure prediction techniques for template-free modelling targets in critical assessment of structure prediction (CASP 9) 4 . Secondary structure, however, is a coarse-grained description of local backbone structure because
A Simple Comparison between Specific Protein Secondary
List of protein structure prediction software Wikipedia
Data Representation Influences Protein Secondary Structure Prediction using Artificial Neural Networks Owen Lamont, Hiew Hong Liang and Matthew Bellgard*
The PSIPRED protein structure prediction server
Protein Structure Prediction Using Neural Networks Martha Mercaldi Kasia Wilamowska Literature Review December 16, 2003. The Protein Folding Problem. Evolution of Neural Networks • Neural networks originally designed to approximate connections between neurons in the brain. Evolution of Neural Networks. Why use Neural Nets for Protein Folding? •Successful applications in: –Secondary
Protein Structure Prediction biostat.wisc.edu
Prediction of protein secondary structure at 80% accuracy
The accuracy of the protein secondary structure prediction programs differs from each other. For molecular biologist, correct structure prediction is a more important factor for understanding protein function, reconstructing protein structures, studying protein–protein
Protein Structure Prediction biostat.wisc.edu
Protein Secondary Structure Prediction Using Cascaded
A Simple Comparison between Specific Protein Secondary
structure of the models predicted by protein structure prediction techniques for template-free modelling targets in critical assessment of structure prediction (CASP 9) 4 . Secondary structure, however, is a coarse-grained description of local backbone structure because
Protein Secondary Structure Prediction Using Cascaded
Sequence Representation and Prediction of Protein
The PSIPRED protein structure prediction server
J . .tlol. Bioi. (1992) 225. W49-1063 . Hybrid System for Protein Secondary Structure Prediction. Xiru Zhang, Jill . P. Mesirov and David L. Waltz
Protein Structure Prediction biostat.wisc.edu
Figure from B. Rost, “Protein Structure in 1D, 2D, and 3D”, The Encyclopaedia of Computational Chemistry, 1998 predicted secondary structure and solvent accessibility known secondary structure (E = beta strand) and solvent accessibility
Data Representation Influences Protein Secondary Structure
A Simple Comparison between Specific Protein Secondary
(Improved prediction of protein secondary structure PNAS
structure of the models predicted by protein structure prediction techniques for template-free modelling targets in critical assessment of structure prediction (CASP 9) 4 . Secondary structure, however, is a coarse-grained description of local backbone structure because
Minimum description length based protein secondary
Prediction of protein secondary structure at 80% accuracy
out of all of the secondary structure prediction methods evaluated, achieving an average Q 3 score of 73.4% over the hardest category and an overall average of 77.3%
PredictProtein Protein Sequence Analysis Prediction of
Minimum description length based protein secondary
This research draws on ideas from related structure prediction fields such as structural class, secondary structure content and protein function prediction, to develop a new, comprehensive and improved representation of protein sequences and to investigate which factors and prediction algorithms result in improved discrimination between the three secondary structures. Separate …
Deep Supervised and Convolutional Generative Stochastic
Sequence Representation and Prediction of Protein
Protein secondary structure prediction (PSSP) is a fundamental task in protein science and computational biology, and it can be used to understand protein 3-dimensional (3-D) structures, further, to learn their biological functions.
Protein Structure Prediction Universitetet i Bergen
(PDF) JPred4 A protein secondary structure prediction server
The present work focuses on secondary structure prediction of proteins. The data mining model is implemented to predict the various parameters related to the secondary structure. These parameters include the alpha helix, beta sheets and hairpin turn. Cluster analysis is used to implement the secondary structure prediction. Key Words: Data mining, Cluster analysis, Protein structure prediction
Minimum description length based protein secondary
The PSIPRED protein structure prediction server
The accuracy of the protein secondary structure prediction programs differs from each other. For molecular biologist, correct structure prediction is a more important factor for understanding protein function, reconstructing protein structures, studying protein–protein
Protein Structure Prediction Universitetet i Bergen
The present work focuses on secondary structure prediction of proteins. The data mining model is implemented to predict the various parameters related to the secondary structure. These parameters include the alpha helix, beta sheets and hairpin turn. Cluster analysis is used to implement the secondary structure prediction. Key Words: Data mining, Cluster analysis, Protein structure prediction
Minimum description length based protein secondary
Deep Supervised and Convolutional Generative Stochastic
This research draws on ideas from related structure prediction fields such as structural class, secondary structure content and protein function prediction, to develop a new, comprehensive and improved representation of protein sequences and to investigate which factors and prediction algorithms result in improved discrimination between the three secondary structures. Separate …
PredictProtein Protein Sequence Analysis Prediction of
Deep Supervised and Convolutional Generative Stochastic Network for Protein Secondary Structure Prediction Jian Zhou JZTHREE@PRINCETON.EDU Olga G. Troyanskaya OGT@CS.
Minimum description length based protein secondary
service for protein structure prediction, protein sequence analysis, protein function prediction, protein sequence alignments, bioinformatics PredictProtein – Protein Sequence Analysis, Prediction of Structural and Functional Features
List of protein structure prediction software Wikipedia
The accuracy of the protein secondary structure prediction programs differs from each other. For molecular biologist, correct structure prediction is a more important factor for understanding protein function, reconstructing protein structures, studying protein–protein
PredictProtein Protein Sequence Analysis Prediction of
A Simple Comparison between Specific Protein Secondary
(PDF) JPred4 A protein secondary structure prediction server
Prediction of Protein Secondary Structure at 80% Accuracy Thomas Nordahl Petersen, 1* Claus Lundegaard,1 Morten Nielsen, Henrik Bohr,2 Jakob Bohr, 2Søren Brunak,
Protein secondary structure prediction A survey of the
(Improved prediction of protein secondary structure PNAS
Data Representation Influences Protein Secondary Structure Prediction using Artificial Neural Networks Owen Lamont, Hiew Hong Liang and Matthew Bellgard*
Sequence Representation and Prediction of Protein
out of all of the secondary structure prediction methods evaluated, achieving an average Q 3 score of 73.4% over the hardest category and an overall average of 77.3%
Minimum description length based protein secondary
List of protein structure prediction software Wikipedia
Data Representation Influences Protein Secondary Structure Prediction using Artificial Neural Networks Owen Lamont, Hiew Hong Liang and Matthew Bellgard*
Sequence Representation and Prediction of Protein
(PDF) The JPred 3 secondary structure prediction server
MOLECULAR MODELING OF PROTEINS AND MATHEMATICAL PREDICTION OF PROTEIN STRUCTURE ARNOLD NEUMAIER ⁄ Abstract. This paper discusses the mathematical formulation of and solution attempts for the
(PDF) JPred4 A protein secondary structure prediction server
Figure from B. Rost, “Protein Structure in 1D, 2D, and 3D”, The Encyclopaedia of Computational Chemistry, 1998 predicted secondary structure and solvent accessibility known secondary structure (E = beta strand) and solvent accessibility
Deep Supervised and Convolutional Generative Stochastic
The probability for a given amino acid to be found in a given secondary structure element is defined based on analyses of its relative frequencies in alpha-helices, beta-sheets and turns in experimentally
(PDF) JPred4 A protein secondary structure prediction server
(PDF) The JPred 3 secondary structure prediction server
Prediction of protein secondary structure at 80% accuracy
Secondary Structure • The primary sequence or main chain of the protein must organize itself to form a compact structure. This is done in an elegant fashion by forming secondary structure elements • The two most common secondary structure elements are alpha helices and beta sheets, formed by repeating amino acids with the same (φ,ψ) angles • There are other secondary structure elements
Protein Structure Prediction biostat.wisc.edu
Improving prediction of secondary structure local
A Simple Comparison between Specific Protein Secondary
JPred4 results summary page (1) with the results of predictions presented in SVG (2). Links to detailed and simple reports in coloured HTML/PS/PDF formats (3).
(Improved prediction of protein secondary structure PNAS
(PDF) The JPred 3 secondary structure prediction server
This list of protein structure prediction software summarizes commonly used software tools in protein structure prediction, including homology modeling, protein threading, ab initio methods, secondary structure prediction, and transmembrane helix and signal peptide prediction.
Data Representation Influences Protein Secondary Structure
(Improved prediction of protein secondary structure PNAS
Figure from B. Rost, “Protein Structure in 1D, 2D, and 3D”, The Encyclopaedia of Computational Chemistry, 1998 predicted secondary structure and solvent accessibility known secondary structure (E = beta strand) and solvent accessibility
Deep Supervised and Convolutional Generative Stochastic
The present work focuses on secondary structure prediction of proteins. The data mining model is implemented to predict the various parameters related to the secondary structure. These parameters include the alpha helix, beta sheets and hairpin turn. Cluster analysis is used to implement the secondary structure prediction. Key Words: Data mining, Cluster analysis, Protein structure prediction
List of protein structure prediction software Wikipedia
Minimum description length based protein secondary
The PSIPRED protein structure prediction server
J . .tlol. Bioi. (1992) 225. W49-1063 . Hybrid System for Protein Secondary Structure Prediction. Xiru Zhang, Jill . P. Mesirov and David L. Waltz
Sequence Representation and Prediction of Protein
Protein secondary structure prediction A survey of the
List of protein structure prediction software Wikipedia
structure of the models predicted by protein structure prediction techniques for template-free modelling targets in critical assessment of structure prediction (CASP 9) 4 . Secondary structure, however, is a coarse-grained description of local backbone structure because
Protein Secondary Structure Prediction Using Cascaded
(Improved prediction of protein secondary structure PNAS
Figure from R. Lathrop et al, “Analysis and Algorithms for Protein Sequence-Structure Alignment” • a threading can be represented as a vector , where each element …
The PSIPRED protein structure prediction server
Improving prediction of secondary structure local
Protein Structure Prediction Using Neural Networks Martha Mercaldi Kasia Wilamowska Literature Review December 16, 2003. The Protein Folding Problem. Evolution of Neural Networks • Neural networks originally designed to approximate connections between neurons in the brain. Evolution of Neural Networks. Why use Neural Nets for Protein Folding? •Successful applications in: –Secondary
A Simple Comparison between Specific Protein Secondary
(PDF) The JPred 3 secondary structure prediction server
(PDF) JPred4 A protein secondary structure prediction server
The present work focuses on secondary structure prediction of proteins. The data mining model is implemented to predict the various parameters related to the secondary structure. These parameters include the alpha helix, beta sheets and hairpin turn. Cluster analysis is used to implement the secondary structure prediction. Key Words: Data mining, Cluster analysis, Protein structure prediction
List of protein structure prediction software Wikipedia
There is no recent blind prediction test for the PROFphd secondary structure prediction algorithm in the PredictProtein secondary structure prediction method, though the earlier PROFsec reported 76% .
Deep Supervised and Convolutional Generative Stochastic
Sequence Representation and Prediction of Protein