Protein secondary structure prediction methods pdf
Nevertheless, many of the secondary-structure prediction methods available today (e.g., in University of Wisconsin Genetics Computer Group (GCG) [88], or from Internet services [89]) are unfortunately still using the dinosaurs of secondary-structure prediction.
Rangwala—Protein Structure Prediction Methods Ja sources of LiMited structuraL data 267 suring the secondary structure content of a protein.
The PSIPRED Protein Sequence Analysis Workbench aggregates several UCL structure prediction methods into one location. Users can submit a protein sequence, perform the predictions of their choice and receive the results of the prediction via e-mail or the web.
1/07/2008 · Secondary structure prediction is an important tool in a structural biologist’s toolbox for the analysis of the significant numbers of proteins, which have no sequence similarity to proteins of known structure. Jpred is a secondary structure prediction server that is a well used and accurate source of predicted secondary structure.
Protein Secondary Structure Prediction Manpreet Singh, Parvinder Singh Sandhu, and Reet Kamal Kaur D World Academy of Science, Engineering and Technology 42 2008 458. in the native states of proteins. This structure class includes regions in the protein of these patterns but it does not include the coordinates of residues. Tertiary Structure is the native state, or folded form, of a single
268 HYBRID METHODS FOR PROTEIN STRUCTURE PREDICTION NMR spectroscopy employs paramagnetic ions to induce the anisotropic pseudo contact shift (PCS) in NMR active nuclei of a protein molecule [10] .
I-TASSER (Iterative Threading ASSEmbly Refinement) is a hierarchical approach to protein structure and function prediction. It first identifies structural templates from the PDB by multiple threading approach LOMETS , with full-length atomic models constructed by iterative template-based fragment assembly simulations.
In this review computational methods for prediction of the protein structure are described and their use towards the drug design is discussed. Citation: Nishant T, Sathish Kumar D, VVL Pavan Kumar A (2011) Computational Methods for Protein Structure Prediction and Its Application in Drug
This paper develops a novel sequence-based method, tetra-peptide-based increment of diversity with quadratic discriminant analysis (TPIDQD for short), for protein secondary-structure prediction. The proposed TPIDQD method is based on tetra-peptide signals and is used to predict the structure of the
Secondary structure prediction is a useful first step in understanding how the amino acid sequence of a protein determines the native state. The most accurate secondary structure prediction algorithms are based on neural networks1, 2 and nearest-neighbor algorithms.7, 27 Methods trained and tested on groups of aligned, homologous sequences, as
better results than any other existing secondary structure prediction method. A long series of similar prediction methods followed, leading to the PHD system [4], the first method to reach the 70% accuracy barrier, and certainly the most successful for many years
JPred Tutorials; How to make a prediction from a single sequence and what the output means (PDF). Submitting more than one sequence to JPred as a batch job (PDF).
The method is implemented in this server based on the descrption in the following paper; Peter Prevelige, Jr. and Gerald D. Fasman (1989) Chapter 9: Chou-Fasman Prediction of the Secondary Structure of Proteins: The Chou-Fasman-Prevelige Algorithm.
As recent successful strategies for secondary structure prediction all rely on multiple sequence information, some methods for accurate protein multiple sequence alignments will also be described. While the main focus is on prediction methods for globular proteins, also the prediction of trans-membrane segments within membrane proteins will be briefly summarised. Finally, an integrated
prediction of the protein secondary structure from the amino acid sequence. The method is based on the most recent version of the standard GOR algorithm. A significant improvement is obtained by combining multiple sequence . International Journal of Computer Applications (0975 – 8887) Volume 73– No.1, July 2013 3 alignments with the GOR method. Additional improvement in the predictions is
Protein Engineering vol.16 no.2 pp.103–107, 2003 DOI: 10.1093/proeng/gzg019 A pentapeptide-based method for protein secondary structure prediction
Protein Structure Prediction an overview ScienceDirect

A Short Review on Protein Secondary Structure Prediction
Protein secondary structure prediction is based on the is weLl kt na g prediction of protein 1-D structure from the sequence of is relatively slow compared to other machine learning aminoacid residues in the target protein [3]. Several methods approaches. In case of protein structure prediction, the have been proposed to find out the secondary structure based problem becomes …
RELIABILITY OF METHODS FOR PREDICTION OF PROTEIN STRUCTURE Prediction of Secondary Structure Prediction of Tertiary Structure Benchmarking a Method of Prediction Blind Automatic Assessments The CASP Experiments AB-INITIO METHODS FOR PREDICTION OF PROTEIN STRUCTURES The Energy of a Protein Configuration Interactions and Energies Covalent …
In secondary structure prediction, the average Q3 accuracy of WDSP in jack-knife test reaches 93.7%. A disease related protein LRRK2 was used as a representive example to demonstrate the structure prediction.
The main aim of this work is practical realization of protein secondary structure prediction method. The created program is supplemented by graphical user interface. In the final part the results of the program based on Chou- Fasman method are compared to …
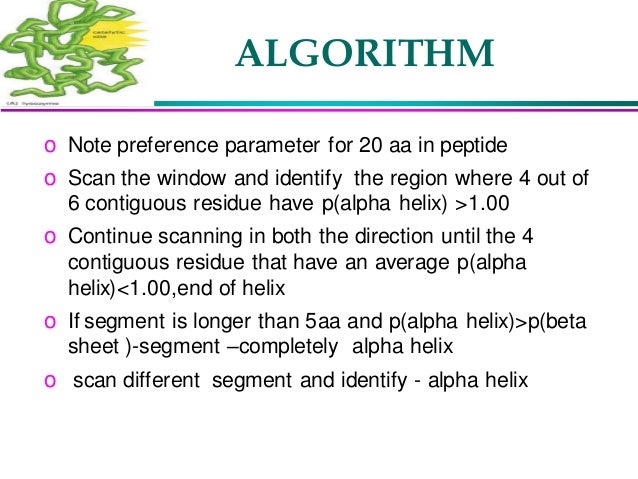
Because of the difficulty of the general protein structure prediction problem, an alternative approach for predicting protein structure is “bottom-up”: here, the goal is to focus on specific, local three-dimensional structures, and develop specialized computational methods
ing prediction methods is overtting. This is the reason why we investigate the implementation This is the reason why we investigate the implementation of Support Vector Machines to perform the task.
in early stages of protein structure prediction • The “Rosetta all-atom energy function,” which depends on the position of every atom, is used in late stages
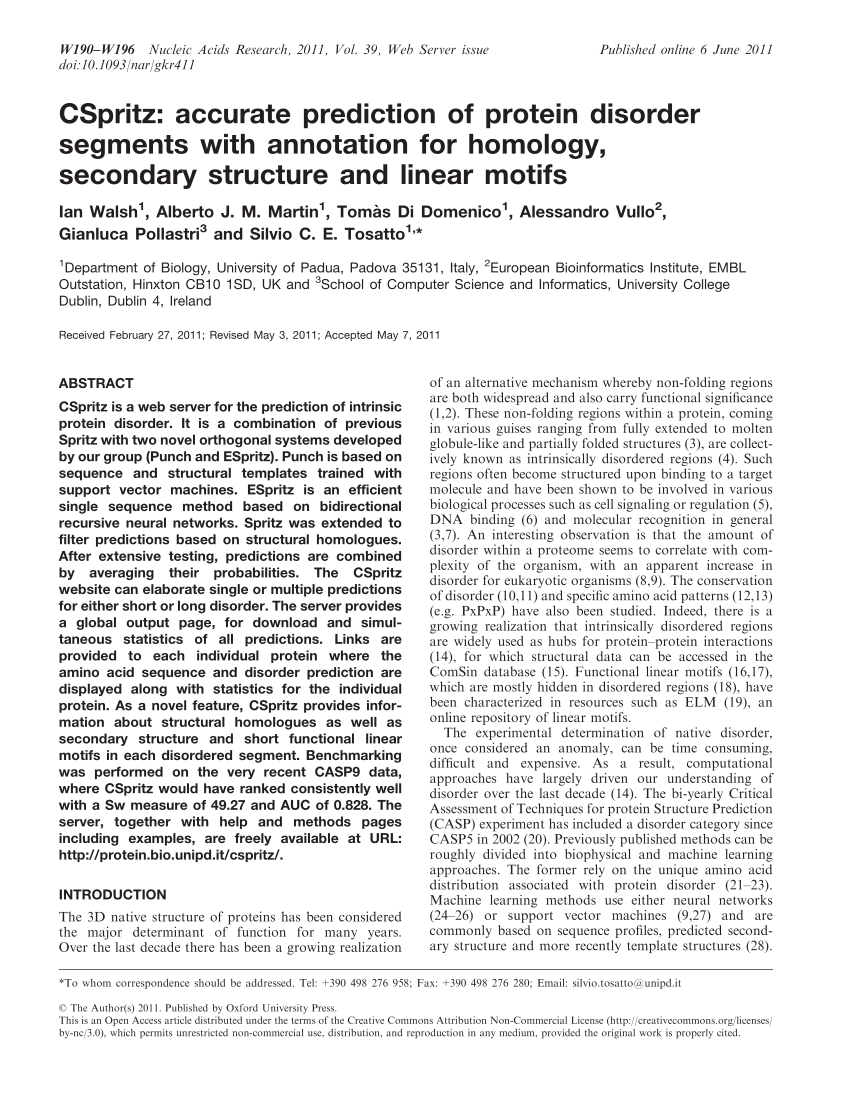
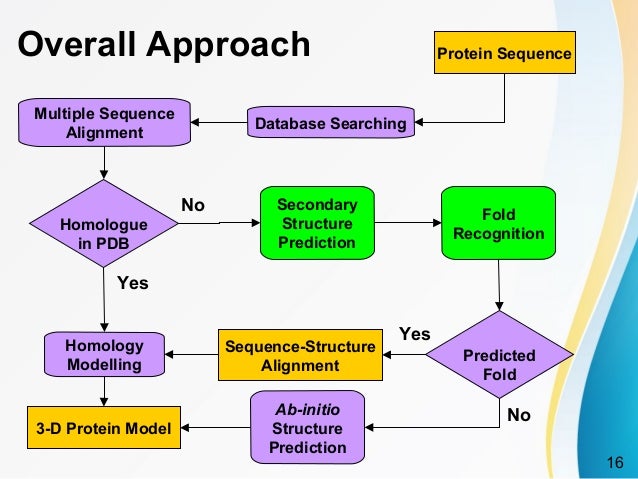
The Chou-Fasman method of secondary structure prediction depends on assigning a set of prediction values to a residue and then applying a simple algorithm to those numbers[3].
• but methods that give us apartial description of the 3D structure are still helpful . 2 Protein Architecture • proteins are polymers consisting of amino acids linked by peptide bonds • each amino acid consists of – a central carbon atom – an amino group – a carboxyl group – a side chain • differences in side chains distinguish different amino acids NH2 COOH Peptide Bonds
Although knowledge of the secondary structure alone is not as useful as a full three-dimensional model, secondary structure predictions provide important constraints for fold-recognition techniques as well as in homology modelling (18,19), ab initio and constraint-based tertiary structure prediction methods . Secondary structure predictions can also help in the identification of functional
Lecture 2 Protein secondary structure prediction Computational Aspects of Molecular Structure Teresa Przytycka, PhD . Assumptions in secondary structure prediction • Goal: classify each residuum as alpha, beta or coil. • Assumption: Secondary structure of a residuum is determined by the amino acid at the given position and amino acids at the neighboring. • Chameleon sequence: A …
Protein Secondary Structure Prediction Based on Position-specific Scoring Matrices David T. Jones Department of Biological Sciences, University of Warwick, Coventry CV4 7AL United Kingdom A two-stage neural network has been used to predict protein secondary structure based on the position specific scoring matrices generated by PSI-BLAST. Despite the simplicity and convenience of the …
One method used to characterize the secondary structure of a protein is circular dichroism spectroscopy (CD). The different types of secondary structure, α-helix, ß-sheet and random coil, all have characteristic circular dichroism spectra in the far-uv region of the spectrum (190-250 nm). These spectra can be used to approximate the fraction of the entire protein made up of each type of
I-TASSER server for protein structure and function prediction
Early secondary structure prediction methods [47] were based on extracting the statistical correlations between a window of consecutive amino acids in a protein sequence and
Batch submission of multiple sequences for individual secondary structure prediction could be done using a file in FASTA format (see link to an example above) and each sequence must be given a unique name (up to 25 characters with no spaces).
This server predicts secondary structure of protein from the amino acid sequence. In this server, Chou & Fasman algorithm has been implemented.
Jenny, T. F. & Benner, S. A. (1994) A prediction of the secondary structure of the pleckstrin homology domain, A prediction of the secondary structure of the pleckstrin homology domain, PROTEINS: Structure, Function and Genetics, 20, 1-3.
Protein Structure Prediction. Protein structure prediction is the method of inference of protein’s 3D structure from its amino acid sequence through the use of computational algorithms.
Ph.D. in Electronic and Computer Engineering Dept. of Electrical and Electronic Engineering University of Cagliari Protein Secondary Structure Prediction: Novel Methods and
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its folding and its secondary and tertiary structure from its primary structure. – crime analysis and prediction using data mining pdf A new dataset of 396 protein domains is developed and used to evaluate the performance of the protein secondary structure prediction algorithms DSC, PHD, NNSSP, and PREDATOR. The maximum theoretical Q 3 accuracy for combination of these methods is shown to be 78%.
Summary: We have created the GOR V web server for protein secondary structure prediction. The GOR V algorithm combines information theory, Bayesian statistics and evolutionary information. In its fifth version, the GOR method reached (with the full jack-knife procedure) an accuracy of prediction Q3 of 73.5%. Although GOR V has been among the most successful methods, its online unavailability
Modern protein secondary structure prediction methods are based on exploiting evolutionary information contained in multiple sequence alignments. Critical steps in the secondary structure prediction process are (i) the selection of a set of sequences that are …
Protein Secondary Structure Prediciton-and-Motif Searching with Scansite. Outline • Brief review of protein structure • Chou-Fasman predictions • Garnier, Osguthorpe and Robson • Helical wheels and hydrophobic moments • Neural networks • Nearest neighbor methods • Consensus prediction approaches . Hierarchy of protein structure . implies planarity Reasonance of peptide bond
Overall, protein secondary structure can be regarded as a bridge that links the primary sequence and tertiary structure and thus, is used by many structure and functional analysis tools 15-18 . Protein SS prediction has been extensively studied 10-12,19-35 .
Protein Structure Analysis Majid Masso Secondary Structure: Computational Problems Secondary structure characterization Secondary structure assignment Protein structure classification Secondary structure prediction. 2 Protein Basic Structure • A protein is made of a chain of amino acids. • There are 20 amino acids found in nature • Each amino acid is coded in the DNA by one or …
Protein Structure Prediction: Secondary Structure Ingo Ruczinski Department of Biostatistics, Johns Hopkins University Protein Folding vs Structure Prediction ¥Protein folding is concerned with the process of the protein taking its three dimensional shape. ¥Protein structure prediction is solely concerned with the 3D structure of the protein. Levinthal Õs Paradox ¥If each amino acid can
Beginning with secondary structure prediction based on sequence only, the book continues by exploring secondary structure prediction based on evolution information, prediction of solvent accessible surface areas and backbone torsion angles, model building, global structural properties, functional properties, as well as visualizing interior and protruding regions in proteins.
Protein structure prediction from amino acid sequence is a fundamental scientific problem and it is regarded as a grand challenge in computational biology and chemistry.
Abstract. While the prediction of a native protein structure from sequence continues to remain a challenging problem, over the past decades computational methods have become quite successful in exploiting the mechanisms behind secondary structure formation.
Prediction of protein secondary structure from the amino acid sequence is a classical bioinformatics problem. Common methods use feed forward neural networks …
[14] It has also been anticipated that the combination of protein secondary structure prediction methods with additional protein structure features has found to provide more accurate results.
Available online at www.sciencedirect.com Computers and Chemical Engineering 32 (2008) 78–88 Prediction of secondary structures of proteins using a two-stage method
sequences of amino acids play in the formation of protein secondary structures. By applying the technique, a performance score of Q 8 = 59 : 2% is achieved, with a cor- …
the secondary structure prediction. In the absence of high-resolution structures, CD is regarded as the method of choice, providing structural information of pro-
JPred4 – is the latest version of the popular JPred protein secondary structure prediction server which provides predictions by the JNet algorithm, one of the most accurate methods for secondary structure prediction. In addition to protein secondary structure, JPred also makes predictions of solvent accessibility and coiled-coil regions. JPred4 features higher accuracy, with a blind three
for protein secondary structure prediction’, Using classifier fusion techniques for protein secondary structure prediction 421 The paper is organized as follows. The general architecture of the proposed meta-classifier have been explained in Section 2. Section 3 describes three different classifier fusion techniques including ordered weighted averaging, Dempster’s combination rule and
New Methods forAccurate Prediction of Protein Secondary Structure John-Marc Chandonia1,2 and Martin Karplus2,3* 1Department of Cellular and Molecular Pharmacology, University of California at San Francisco, San Francisco, California
Introduction. For protein sequences of unknown structure and/or biological function, one of the first and quite insightful analyses of the linear sequence of amino acids (i.e., the primary structure) is a prediction of the secondary structure.
Hybrid MetHods for Protein structure Prediction ANU
COS551: 11/16/2000 2 1.1 The Chou-Fasman method If you were asked to determine whether an amino acid in a protein of interest is part of a -helix or -sheet, you might think to look in a protein …
A Hybrid Method for Protein Secondary Structure Prediction Shing-Hwang Doong Chi-Yuan Yeh Department of Information Management, ShuTe University
Protein secondary structure prediction is defined as the set of techniques and algorithms in bioinformatics that aim to predict the local secondary structures based only on the knowledge of their primary structure.
curate methods for secondary structure prediction. In addition to protein secondary structure, JPred also makes predictions of solvent accessibility and coiled-coil regions. The JPred service runs up to 94 000 jobs per month and has carried out over 1.5 mil-lion predictions in total for users in 179 countries. The JPred4 web server has been re-implemented in the Bootstrap framework and
Now if the results of secondary structure content prediction by some method for a given protein are α = 40% and β = 0%, while its corresponding observed values are actually α = 60% and β = 5%, this means there is a deviation of60 – 40%| =20% = 0.2 and5 – 0%| = 5% = 0.05 in the prediction of α-helix and β-sheet content, respectively. Such a deviation would represent a significant
Review: Protein Secondary Structure Prediction Continues to Rise Burkhard Rost CUBIC, Department of Biochemistry and Molecular Biophysics, Columbia University, 630 West 168th Street, New York, New York 10032 Received November 20, 2000, and in revised form February 21, 2001; published online June 13, 2001 Methods predicting protein secondary structure improved …
13 Use of Class Prediction to Improve Protein Secondary Structure Prediction Joint Prediction with Methods Based on Sequence Homology Gilbert De/eage and Bernard Roux
Protein Structure Prediction Methods And Protocol PDF

JPred A Protein Secondary Structure Prediction Server
1 Secondary structure prediction
A Hybrid Method for Protein Secondary Structure Prediction
Use of tetrapeptide signals for protein secondary

Protein Secondary Structure Prediction Novel Methods and
https://en.m.wikipedia.org/wiki/List_of_protein_secondary_structure_prediction_programs
Protein Structure Prediction Biostatistics – Departments
bounty hunter tracker 2d 707 manual – A method for WD40 repeat detection and secondary structure
A STUDY OF INTELLIGENT TECHNIQUES FOR PROTEIN SECONDARY

Jeeva Enterprise Grid Enabled Protein Secondary Structure
Computational Methods for Protein Secondary Structure
Integrating Protein Secondary Structure Prediction and
Jeeva Enterprise Grid Enabled Protein Secondary Structure
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its folding and its secondary and tertiary structure from its primary structure.
• but methods that give us apartial description of the 3D structure are still helpful . 2 Protein Architecture • proteins are polymers consisting of amino acids linked by peptide bonds • each amino acid consists of – a central carbon atom – an amino group – a carboxyl group – a side chain • differences in side chains distinguish different amino acids NH2 COOH Peptide Bonds
This paper develops a novel sequence-based method, tetra-peptide-based increment of diversity with quadratic discriminant analysis (TPIDQD for short), for protein secondary-structure prediction. The proposed TPIDQD method is based on tetra-peptide signals and is used to predict the structure of the
In secondary structure prediction, the average Q3 accuracy of WDSP in jack-knife test reaches 93.7%. A disease related protein LRRK2 was used as a representive example to demonstrate the structure prediction.
Using classifier fusion techniques for protein secondary
A Short Review on Protein Secondary Structure Prediction
Jenny, T. F. & Benner, S. A. (1994) A prediction of the secondary structure of the pleckstrin homology domain, A prediction of the secondary structure of the pleckstrin homology domain, PROTEINS: Structure, Function and Genetics, 20, 1-3.
The Chou-Fasman method of secondary structure prediction depends on assigning a set of prediction values to a residue and then applying a simple algorithm to those numbers[3].
Introduction. For protein sequences of unknown structure and/or biological function, one of the first and quite insightful analyses of the linear sequence of amino acids (i.e., the primary structure) is a prediction of the secondary structure.
Beginning with secondary structure prediction based on sequence only, the book continues by exploring secondary structure prediction based on evolution information, prediction of solvent accessible surface areas and backbone torsion angles, model building, global structural properties, functional properties, as well as visualizing interior and protruding regions in proteins.
Protein Secondary Structure Prediciton-and-Motif Searching with Scansite. Outline • Brief review of protein structure • Chou-Fasman predictions • Garnier, Osguthorpe and Robson • Helical wheels and hydrophobic moments • Neural networks • Nearest neighbor methods • Consensus prediction approaches . Hierarchy of protein structure . implies planarity Reasonance of peptide bond
Because of the difficulty of the general protein structure prediction problem, an alternative approach for predicting protein structure is “bottom-up”: here, the goal is to focus on specific, local three-dimensional structures, and develop specialized computational methods
prediction of the protein secondary structure from the amino acid sequence. The method is based on the most recent version of the standard GOR algorithm. A significant improvement is obtained by combining multiple sequence . International Journal of Computer Applications (0975 – 8887) Volume 73– No.1, July 2013 3 alignments with the GOR method. Additional improvement in the predictions is
In secondary structure prediction, the average Q3 accuracy of WDSP in jack-knife test reaches 93.7%. A disease related protein LRRK2 was used as a representive example to demonstrate the structure prediction.
COS551: 11/16/2000 2 1.1 The Chou-Fasman method If you were asked to determine whether an amino acid in a protein of interest is part of a -helix or -sheet, you might think to look in a protein …
1/07/2008 · Secondary structure prediction is an important tool in a structural biologist’s toolbox for the analysis of the significant numbers of proteins, which have no sequence similarity to proteins of known structure. Jpred is a secondary structure prediction server that is a well used and accurate source of predicted secondary structure.
New Methods forAccurate Prediction of Protein Secondary Structure John-Marc Chandonia1,2 and Martin Karplus2,3* 1Department of Cellular and Molecular Pharmacology, University of California at San Francisco, San Francisco, California
A Hybrid Method for Protein Secondary Structure Prediction Shing-Hwang Doong Chi-Yuan Yeh Department of Information Management, ShuTe University
Ph.D. in Electronic and Computer Engineering Dept. of Electrical and Electronic Engineering University of Cagliari Protein Secondary Structure Prediction: Novel Methods and
Lecture 2 Protein secondary structure prediction Computational Aspects of Molecular Structure Teresa Przytycka, PhD . Assumptions in secondary structure prediction • Goal: classify each residuum as alpha, beta or coil. • Assumption: Secondary structure of a residuum is determined by the amino acid at the given position and amino acids at the neighboring. • Chameleon sequence: A …
A Short Review on Protein Secondary Structure Prediction
Prediction of protein secondary structure content
This paper develops a novel sequence-based method, tetra-peptide-based increment of diversity with quadratic discriminant analysis (TPIDQD for short), for protein secondary-structure prediction. The proposed TPIDQD method is based on tetra-peptide signals and is used to predict the structure of the
268 HYBRID METHODS FOR PROTEIN STRUCTURE PREDICTION NMR spectroscopy employs paramagnetic ions to induce the anisotropic pseudo contact shift (PCS) in NMR active nuclei of a protein molecule [10] .
Beginning with secondary structure prediction based on sequence only, the book continues by exploring secondary structure prediction based on evolution information, prediction of solvent accessible surface areas and backbone torsion angles, model building, global structural properties, functional properties, as well as visualizing interior and protruding regions in proteins.
in early stages of protein structure prediction • The “Rosetta all-atom energy function,” which depends on the position of every atom, is used in late stages
Summary: We have created the GOR V web server for protein secondary structure prediction. The GOR V algorithm combines information theory, Bayesian statistics and evolutionary information. In its fifth version, the GOR method reached (with the full jack-knife procedure) an accuracy of prediction Q3 of 73.5%. Although GOR V has been among the most successful methods, its online unavailability
better results than any other existing secondary structure prediction method. A long series of similar prediction methods followed, leading to the PHD system [4], the first method to reach the 70% accuracy barrier, and certainly the most successful for many years
Although knowledge of the secondary structure alone is not as useful as a full three-dimensional model, secondary structure predictions provide important constraints for fold-recognition techniques as well as in homology modelling (18,19), ab initio and constraint-based tertiary structure prediction methods . Secondary structure predictions can also help in the identification of functional
A new dataset of 396 protein domains is developed and used to evaluate the performance of the protein secondary structure prediction algorithms DSC, PHD, NNSSP, and PREDATOR. The maximum theoretical Q 3 accuracy for combination of these methods is shown to be 78%.
13 Use of Class Prediction to Improve Protein Secondary Structure Prediction Joint Prediction with Methods Based on Sequence Homology Gilbert De/eage and Bernard Roux
The main aim of this work is practical realization of protein secondary structure prediction method. The created program is supplemented by graphical user interface. In the final part the results of the program based on Chou- Fasman method are compared to …
Prediction of protein secondary structure from the amino acid sequence is a classical bioinformatics problem. Common methods use feed forward neural networks …
The PSIPRED Protein Sequence Analysis Workbench aggregates several UCL structure prediction methods into one location. Users can submit a protein sequence, perform the predictions of their choice and receive the results of the prediction via e-mail or the web.
Batch submission of multiple sequences for individual secondary structure prediction could be done using a file in FASTA format (see link to an example above) and each sequence must be given a unique name (up to 25 characters with no spaces).
Protein Structure Prediction: Secondary Structure Ingo Ruczinski Department of Biostatistics, Johns Hopkins University Protein Folding vs Structure Prediction ¥Protein folding is concerned with the process of the protein taking its three dimensional shape. ¥Protein structure prediction is solely concerned with the 3D structure of the protein. Levinthal Õs Paradox ¥If each amino acid can
Protein Structure Prediction Methods And Protocol PDF
Protein Secondary Structure Prediction CiteSeerX
Protein Secondary Structure Prediction Manpreet Singh, Parvinder Singh Sandhu, and Reet Kamal Kaur D World Academy of Science, Engineering and Technology 42 2008 458. in the native states of proteins. This structure class includes regions in the protein of these patterns but it does not include the coordinates of residues. Tertiary Structure is the native state, or folded form, of a single
• but methods that give us apartial description of the 3D structure are still helpful . 2 Protein Architecture • proteins are polymers consisting of amino acids linked by peptide bonds • each amino acid consists of – a central carbon atom – an amino group – a carboxyl group – a side chain • differences in side chains distinguish different amino acids NH2 COOH Peptide Bonds
Protein Structure Prediction: Secondary Structure Ingo Ruczinski Department of Biostatistics, Johns Hopkins University Protein Folding vs Structure Prediction ¥Protein folding is concerned with the process of the protein taking its three dimensional shape. ¥Protein structure prediction is solely concerned with the 3D structure of the protein. Levinthal Õs Paradox ¥If each amino acid can
Batch submission of multiple sequences for individual secondary structure prediction could be done using a file in FASTA format (see link to an example above) and each sequence must be given a unique name (up to 25 characters with no spaces).
Beginning with secondary structure prediction based on sequence only, the book continues by exploring secondary structure prediction based on evolution information, prediction of solvent accessible surface areas and backbone torsion angles, model building, global structural properties, functional properties, as well as visualizing interior and protruding regions in proteins.
the secondary structure prediction. In the absence of high-resolution structures, CD is regarded as the method of choice, providing structural information of pro-
Protein Structure Prediction. Protein structure prediction is the method of inference of protein’s 3D structure from its amino acid sequence through the use of computational algorithms.
ing prediction methods is overtting. This is the reason why we investigate the implementation This is the reason why we investigate the implementation of Support Vector Machines to perform the task.
sequences of amino acids play in the formation of protein secondary structures. By applying the technique, a performance score of Q 8 = 59 : 2% is achieved, with a cor- …
Abstract. While the prediction of a native protein structure from sequence continues to remain a challenging problem, over the past decades computational methods have become quite successful in exploiting the mechanisms behind secondary structure formation.
In secondary structure prediction, the average Q3 accuracy of WDSP in jack-knife test reaches 93.7%. A disease related protein LRRK2 was used as a representive example to demonstrate the structure prediction.
Modern protein secondary structure prediction methods are based on exploiting evolutionary information contained in multiple sequence alignments. Critical steps in the secondary structure prediction process are (i) the selection of a set of sequences that are …
Protein Secondary Structure Prediction
Protein Structure Prediction Biostatistics – Departments
ing prediction methods is overtting. This is the reason why we investigate the implementation This is the reason why we investigate the implementation of Support Vector Machines to perform the task.
Modern protein secondary structure prediction methods are based on exploiting evolutionary information contained in multiple sequence alignments. Critical steps in the secondary structure prediction process are (i) the selection of a set of sequences that are …
A new dataset of 396 protein domains is developed and used to evaluate the performance of the protein secondary structure prediction algorithms DSC, PHD, NNSSP, and PREDATOR. The maximum theoretical Q 3 accuracy for combination of these methods is shown to be 78%.
268 HYBRID METHODS FOR PROTEIN STRUCTURE PREDICTION NMR spectroscopy employs paramagnetic ions to induce the anisotropic pseudo contact shift (PCS) in NMR active nuclei of a protein molecule [10] .
Jenny, T. F. & Benner, S. A. (1994) A prediction of the secondary structure of the pleckstrin homology domain, A prediction of the secondary structure of the pleckstrin homology domain, PROTEINS: Structure, Function and Genetics, 20, 1-3.
Batch submission of multiple sequences for individual secondary structure prediction could be done using a file in FASTA format (see link to an example above) and each sequence must be given a unique name (up to 25 characters with no spaces).
prediction of the protein secondary structure from the amino acid sequence. The method is based on the most recent version of the standard GOR algorithm. A significant improvement is obtained by combining multiple sequence . International Journal of Computer Applications (0975 – 8887) Volume 73– No.1, July 2013 3 alignments with the GOR method. Additional improvement in the predictions is
Using Cluster Analysis for Protein Secondary Structure
Prediction of Protein Secondary Structure Yaoqi Zhou
Secondary structure prediction is a useful first step in understanding how the amino acid sequence of a protein determines the native state. The most accurate secondary structure prediction algorithms are based on neural networks1, 2 and nearest-neighbor algorithms.7, 27 Methods trained and tested on groups of aligned, homologous sequences, as
for protein secondary structure prediction’, Using classifier fusion techniques for protein secondary structure prediction 421 The paper is organized as follows. The general architecture of the proposed meta-classifier have been explained in Section 2. Section 3 describes three different classifier fusion techniques including ordered weighted averaging, Dempster’s combination rule and
In this review computational methods for prediction of the protein structure are described and their use towards the drug design is discussed. Citation: Nishant T, Sathish Kumar D, VVL Pavan Kumar A (2011) Computational Methods for Protein Structure Prediction and Its Application in Drug
Protein secondary structure prediction is defined as the set of techniques and algorithms in bioinformatics that aim to predict the local secondary structures based only on the knowledge of their primary structure.
The Chou-Fasman method of secondary structure prediction depends on assigning a set of prediction values to a residue and then applying a simple algorithm to those numbers[3].
ing prediction methods is overtting. This is the reason why we investigate the implementation This is the reason why we investigate the implementation of Support Vector Machines to perform the task.
JPred4 a protein secondary structure prediction server
Methods for prediction of secondary structure in proteins
In this review computational methods for prediction of the protein structure are described and their use towards the drug design is discussed. Citation: Nishant T, Sathish Kumar D, VVL Pavan Kumar A (2011) Computational Methods for Protein Structure Prediction and Its Application in Drug
Protein Structure Prediction. Protein structure prediction is the method of inference of protein’s 3D structure from its amino acid sequence through the use of computational algorithms.
Abstract. While the prediction of a native protein structure from sequence continues to remain a challenging problem, over the past decades computational methods have become quite successful in exploiting the mechanisms behind secondary structure formation.
The PSIPRED Protein Sequence Analysis Workbench aggregates several UCL structure prediction methods into one location. Users can submit a protein sequence, perform the predictions of their choice and receive the results of the prediction via e-mail or the web.
Batch submission of multiple sequences for individual secondary structure prediction could be done using a file in FASTA format (see link to an example above) and each sequence must be given a unique name (up to 25 characters with no spaces).
As recent successful strategies for secondary structure prediction all rely on multiple sequence information, some methods for accurate protein multiple sequence alignments will also be described. While the main focus is on prediction methods for globular proteins, also the prediction of trans-membrane segments within membrane proteins will be briefly summarised. Finally, an integrated
ing prediction methods is overtting. This is the reason why we investigate the implementation This is the reason why we investigate the implementation of Support Vector Machines to perform the task.
prediction of the protein secondary structure from the amino acid sequence. The method is based on the most recent version of the standard GOR algorithm. A significant improvement is obtained by combining multiple sequence . International Journal of Computer Applications (0975 – 8887) Volume 73– No.1, July 2013 3 alignments with the GOR method. Additional improvement in the predictions is
JPred Tutorials; How to make a prediction from a single sequence and what the output means (PDF). Submitting more than one sequence to JPred as a batch job (PDF).
Protein Secondary Structure Prediction Novel Methods and
Methods for prediction of secondary structure in proteins
A Hybrid Method for Protein Secondary Structure Prediction Shing-Hwang Doong Chi-Yuan Yeh Department of Information Management, ShuTe University
I-TASSER server for protein structure and function prediction
Secondary Structure Prediction methods and links
This paper develops a novel sequence-based method, tetra-peptide-based increment of diversity with quadratic discriminant analysis (TPIDQD for short), for protein secondary-structure prediction. The proposed TPIDQD method is based on tetra-peptide signals and is used to predict the structure of the
Proteins Structure Function and Bioinformatics
This server predicts secondary structure of protein from the amino acid sequence. In this server, Chou & Fasman algorithm has been implemented.
A Short Review on Protein Secondary Structure Prediction
• but methods that give us apartial description of the 3D structure are still helpful . 2 Protein Architecture • proteins are polymers consisting of amino acids linked by peptide bonds • each amino acid consists of – a central carbon atom – an amino group – a carboxyl group – a side chain • differences in side chains distinguish different amino acids NH2 COOH Peptide Bonds
Proteins Structure Function and Bioinformatics
[14] It has also been anticipated that the combination of protein secondary structure prediction methods with additional protein structure features has found to provide more accurate results.
Prediction of protein secondary structure content
Protein Secondary Structure Prediction
Ph.D. in Electronic and Computer Engineering Dept. of Electrical and Electronic Engineering University of Cagliari Protein Secondary Structure Prediction: Novel Methods and
Secondary Structure Prediction methods and links
Prediction of protein secondary structure content
• but methods that give us apartial description of the 3D structure are still helpful . 2 Protein Architecture • proteins are polymers consisting of amino acids linked by peptide bonds • each amino acid consists of – a central carbon atom – an amino group – a carboxyl group – a side chain • differences in side chains distinguish different amino acids NH2 COOH Peptide Bonds
Protein Secondary Structure Prediction CiteSeerX
Review Protein Secondary Structure Prediction Continues
Secondary Structure Prediction by Chou-Fasman GOR and
The method is implemented in this server based on the descrption in the following paper; Peter Prevelige, Jr. and Gerald D. Fasman (1989) Chapter 9: Chou-Fasman Prediction of the Secondary Structure of Proteins: The Chou-Fasman-Prevelige Algorithm.
Online Analysis Tools Protein Secondary Structure
Secondary Structure Prediction by Chou-Fasman GOR and
In secondary structure prediction, the average Q3 accuracy of WDSP in jack-knife test reaches 93.7%. A disease related protein LRRK2 was used as a representive example to demonstrate the structure prediction.
Proteins Structure Function and Bioinformatics
Computational Methods in Protein Structure Prediction
curate methods for secondary structure prediction. In addition to protein secondary structure, JPred also makes predictions of solvent accessibility and coiled-coil regions. The JPred service runs up to 94 000 jobs per month and has carried out over 1.5 mil-lion predictions in total for users in 179 countries. The JPred4 web server has been re-implemented in the Bootstrap framework and
Protein structure prediction Wikipedia
Protein Secondary Structure Prediction CiteSeerX
sequences of amino acids play in the formation of protein secondary structures. By applying the technique, a performance score of Q 8 = 59 : 2% is achieved, with a cor- …
GOR Method for Protein Structure Prediction using Cluster
Protein Secondary Structure Prediction SpringerLink
Proteins Structure Function and Bioinformatics
The Chou-Fasman method of secondary structure prediction depends on assigning a set of prediction values to a residue and then applying a simple algorithm to those numbers[3].
Prediction of Protein Secondary Structure Yaoqi Zhou
Use of tetrapeptide signals for protein secondary
Proteins Structure Function and Bioinformatics
[14] It has also been anticipated that the combination of protein secondary structure prediction methods with additional protein structure features has found to provide more accurate results.
Computational Methods in Protein Structure Prediction
Protein Structure Analysis School of Systems Biology
Protein structure prediction Wikipedia
Introduction. For protein sequences of unknown structure and/or biological function, one of the first and quite insightful analyses of the linear sequence of amino acids (i.e., the primary structure) is a prediction of the secondary structure.
Protein Structure Prediction Biostatistics – Departments
1/07/2008 · Secondary structure prediction is an important tool in a structural biologist’s toolbox for the analysis of the significant numbers of proteins, which have no sequence similarity to proteins of known structure. Jpred is a secondary structure prediction server that is a well used and accurate source of predicted secondary structure.
GOR Method for Protein Structure Prediction using Cluster
Using classifier fusion techniques for protein secondary
Protein Secondary Structure Prediction Based on Position-specific Scoring Matrices David T. Jones Department of Biological Sciences, University of Warwick, Coventry CV4 7AL United Kingdom A two-stage neural network has been used to predict protein secondary structure based on the position specific scoring matrices generated by PSI-BLAST. Despite the simplicity and convenience of the …
The Jpred 3 secondary structure prediction server
Overall, protein secondary structure can be regarded as a bridge that links the primary sequence and tertiary structure and thus, is used by many structure and functional analysis tools 15-18 . Protein SS prediction has been extensively studied 10-12,19-35 .
Protein Secondary Structure Prediction Novel Methods and
A STUDY OF INTELLIGENT TECHNIQUES FOR PROTEIN SECONDARY
This server predicts secondary structure of protein from the amino acid sequence. In this server, Chou & Fasman algorithm has been implemented.
HYBRID METHODS FOR PROTEIN STRUCTURE PREDICTION
A method for WD40 repeat detection and secondary structure
Protein Structure Prediction Methods And Protocol PDF
better results than any other existing secondary structure prediction method. A long series of similar prediction methods followed, leading to the PHD system [4], the first method to reach the 70% accuracy barrier, and certainly the most successful for many years
Protein Secondary Structure Prediction SpringerLink
Protein Secondary Structure Prediction CiteSeerX
Combining protein secondary structure prediction models
This server predicts secondary structure of protein from the amino acid sequence. In this server, Chou & Fasman algorithm has been implemented.
Computational Methods for Protein Secondary Structure
Combining protein secondary structure prediction models
This server predicts secondary structure of protein from the amino acid sequence. In this server, Chou & Fasman algorithm has been implemented.
PSIPRED protein structure prediction
One method used to characterize the secondary structure of a protein is circular dichroism spectroscopy (CD). The different types of secondary structure, α-helix, ß-sheet and random coil, all have characteristic circular dichroism spectra in the far-uv region of the spectrum (190-250 nm). These spectra can be used to approximate the fraction of the entire protein made up of each type of
Protein structure prediction Wikipedia
ing prediction methods is overtting. This is the reason why we investigate the implementation This is the reason why we investigate the implementation of Support Vector Machines to perform the task.
Protein Secondary Structure Prediction SpringerLink
Protein Structure Prediction an overview ScienceDirect
ing prediction methods is overtting. This is the reason why we investigate the implementation This is the reason why we investigate the implementation of Support Vector Machines to perform the task.
Jeeva Enterprise Grid Enabled Protein Secondary Structure
In this review computational methods for prediction of the protein structure are described and their use towards the drug design is discussed. Citation: Nishant T, Sathish Kumar D, VVL Pavan Kumar A (2011) Computational Methods for Protein Structure Prediction and Its Application in Drug
IEEE REVIEWS IN BIOMEDICAL ENGINEERING VOL. 1 2008 41
HYBRID METHODS FOR PROTEIN STRUCTURE PREDICTION
Jeeva Enterprise Grid Enabled Protein Secondary Structure
The PSIPRED Protein Sequence Analysis Workbench aggregates several UCL structure prediction methods into one location. Users can submit a protein sequence, perform the predictions of their choice and receive the results of the prediction via e-mail or the web.
Prediction of secondary structures of proteins using a two
The Jpred 3 secondary structure prediction server
A method for WD40 repeat detection and secondary structure
Lecture 2 Protein secondary structure prediction Computational Aspects of Molecular Structure Teresa Przytycka, PhD . Assumptions in secondary structure prediction • Goal: classify each residuum as alpha, beta or coil. • Assumption: Secondary structure of a residuum is determined by the amino acid at the given position and amino acids at the neighboring. • Chameleon sequence: A …
PSIPRED protein structure prediction
Protein structure prediction Wikipedia
Lecture 2 Protein secondary structure prediction Computational Aspects of Molecular Structure Teresa Przytycka, PhD . Assumptions in secondary structure prediction • Goal: classify each residuum as alpha, beta or coil. • Assumption: Secondary structure of a residuum is determined by the amino acid at the given position and amino acids at the neighboring. • Chameleon sequence: A …
ExPASy SIB Bioinformatics Resource Portal Resources
PSIPRED protein structure prediction
Secondary Structure Prediction methods and links
Protein Structure Analysis Majid Masso Secondary Structure: Computational Problems Secondary structure characterization Secondary structure assignment Protein structure classification Secondary structure prediction. 2 Protein Basic Structure • A protein is made of a chain of amino acids. • There are 20 amino acids found in nature • Each amino acid is coded in the DNA by one or …
Protein Secondary Structure Prediction CiteSeerX
Combining protein secondary structure prediction models
for protein secondary structure prediction’, Using classifier fusion techniques for protein secondary structure prediction 421 The paper is organized as follows. The general architecture of the proposed meta-classifier have been explained in Section 2. Section 3 describes three different classifier fusion techniques including ordered weighted averaging, Dempster’s combination rule and
New Methods forAccurate Prediction of Protein Secondary
Protein Secondary Structure Prediction CiteSeerX
Computational Methods for Protein Structure Prediction and
The main aim of this work is practical realization of protein secondary structure prediction method. The created program is supplemented by graphical user interface. In the final part the results of the program based on Chou- Fasman method are compared to …
Secondary Structure Prediction methods and links
Jenny, T. F. & Benner, S. A. (1994) A prediction of the secondary structure of the pleckstrin homology domain, A prediction of the secondary structure of the pleckstrin homology domain, PROTEINS: Structure, Function and Genetics, 20, 1-3.
1 Secondary structure prediction
The main aim of this work is practical realization of protein secondary structure prediction method. The created program is supplemented by graphical user interface. In the final part the results of the program based on Chou- Fasman method are compared to …
Use of Class Prediction to Improve Protein Secondary
A STUDY OF INTELLIGENT TECHNIQUES FOR PROTEIN SECONDARY
Prediction of Protein Secondary Structure Yaoqi Zhou
prediction of the protein secondary structure from the amino acid sequence. The method is based on the most recent version of the standard GOR algorithm. A significant improvement is obtained by combining multiple sequence . International Journal of Computer Applications (0975 – 8887) Volume 73– No.1, July 2013 3 alignments with the GOR method. Additional improvement in the predictions is
HYBRID METHODS FOR PROTEIN STRUCTURE PREDICTION
Computational Methods for Protein Structure Prediction and
Protein Secondary Structure Prediction Manpreet Singh, Parvinder Singh Sandhu, and Reet Kamal Kaur D World Academy of Science, Engineering and Technology 42 2008 458. in the native states of proteins. This structure class includes regions in the protein of these patterns but it does not include the coordinates of residues. Tertiary Structure is the native state, or folded form, of a single
Online Analysis Tools Protein Secondary Structure
I-TASSER server for protein structure and function prediction
sequences of amino acids play in the formation of protein secondary structures. By applying the technique, a performance score of Q 8 = 59 : 2% is achieved, with a cor- …
A Short Review on Protein Secondary Structure Prediction
Hybrid MetHods for Protein structure Prediction ANU
the secondary structure prediction. In the absence of high-resolution structures, CD is regarded as the method of choice, providing structural information of pro-
PROTEIN SECONDARY STRUCTURE PREDICTION USING AMINO
In secondary structure prediction, the average Q3 accuracy of WDSP in jack-knife test reaches 93.7%. A disease related protein LRRK2 was used as a representive example to demonstrate the structure prediction.
Integrating Protein Secondary Structure Prediction and