Gene prediction in prokaryotes and eukaryotes pdf
Gene prediction in eukaryotes is notoriously difficult; it is not even a straightforward business to decide how to count and, in humans at least, there is considerable discrepancy between different gene prediction methods . Clearly, phenomena such as alternative splicing and post-translational modifications of genes must also be taken into account. Accurate prediction of alternative gene
In eukaryotes, a gene is a combination of coding segments (exons) that are interrupted by non-coding segments (introns) This makes computational gene prediction in eukaryotes even more
RNA Polymerase: Prokaryotes have one RNA polymerase that handles all transcription while eukaryotes have 3. The prokaryotic RNA polymerase is much …
eukaryotes and between eukaryotes and prokaryotes, with the present genome data these examples will not have a significant role for the prediction of functional interactions between proteins. It is possible that with the sequencing of more species such as the nematodes or yeast species in which neighboring genes do have an above-average probability of being co-regulated, the importance of gene
unicellular eukaryotes (1, 2). Also, with the exception of deterio- rating genomes of some parasitic bacteria, the prokaryotic genomes are highly compact, with densely packed protein-coding genes and a low fraction of noncoding sequences (3). The small genome size is thought to be selected for fast re plication, whereas the high gene density additionally facilitates coregulation of gene
transcription initiation!in plants [12, 20S21].Wefacethesituationthatspecific! promoter!characteristics!that!often!used!in!development!of!promoter!predictors!in!
Evaluation of gene prediction methods for prokaryotes Master’s thesis Stian Engebretsen 15th August 2012. Abstract There are currently over 2,000 completed prokaryotic sequences available. In these DNA sequences the genes lay hidden. The genes codes for proteins that forms the foundation for all organisms. Several programs that try to locate these genes have been developed. However, little
In this unit, the GeneMark and GeneMark.hmm programs are presented as two different methods for the in silico prediction of genes in prokaryotes.
1/07/2005 · The website provides interfaces to the GeneMark family of programs designed and tuned for gene prediction in prokaryotic, eukaryotic and viral genomic sequences. Currently, the server allows the analysis of nearly 200 prokaryotic and >10 eukaryotic genomes using species-specific versions of the software and pre-computed gene models. In addition, genes in prokaryotic sequences from novel
Eukaryotic vs. Prokaryotic Gene Prediction Prokaryotes: – Overall small genome size – High density of CDSs – Low numbers of repeated noncoding sequences – Regulatory regions are close to the protein coding sequences – CDS regions are short Eukaryotes – Opposite of Prokaryotes . Different types of algorithms for gene prediction 1. Ab initio a. Identify genes based on intrinsic factors
Two Approaches to Gene Prediction An Introduction to Bioinformatics Algorithms www.bioalgorithms.info If you could compare the day’s news in English, side-by-side
A promoter is a regulatory region of DNA located upstream (towards the 5′ region) of of a gene, providing a control point for regulated gene transcription. The promoter contains specific DNA sequences that are recognized by proteins known as transcription factors.
GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses John Besemer1 and Mark Borodovsky1,2
Hot, Toxic Eukaryote. Unusually, the single-celled eukaryote red alga, Galdieria sulphuraria, can thrive in hot, acidic springs. This organism is endowed with extraordinary metabolic talents and can consume a variety of strange carbohydrates, as well as turn on photosynthesis when the food runs out.
System I was apparently ancestral to all eukaryotes and was functionally replaced by the HCCS (which is absent in prokaryotes), but whether this happened once or multiple times is an unresolved debate, the implications of which have been hypothesized to apply to rooting the tree of eukaryotes
Genome Analysis Inversions and the dynamics of eukaryotic

AUGUSTUS a web server for gene prediction in eukaryotes
gene structure prediction and annotation is the fundamental step towards the understanding of genome function. A A large number of gene prediction tool and pipeline have …
Introduction Gene Prediction. Gene prediction is the process of identifying specific regions of genomic DNA that encode for genes. A gene is the entire DNA sequence necessary for production of a functional polypeptide or RNA.
– In this paper we propose a novel gene prediction program for prokaryotic sequences. This program has been developed in PERL. The web inter-face uses CGI and front end for data input and viewing the result has also been developed. The open reading
Gene Prediction Ppt – Download as Powerpoint Presentation (.ppt), PDF File (.pdf), Text File (.txt) or view presentation slides online. Scribd is the world’s largest social reading and publishing site.
Background. As more and more genomes are being sequenced, an overview of their genomic features and annotation of their functional elements, which control the expression of each gene or transcription unit of the genome, is a fundamental challenge in genomics and bioinformatics.
Contains fulltext : 30111_compgeofe.pdf (publisher’s version ) (Open Access)This thesis focuses on developing comparative genomics methods in eukaryotes, with an emphasis on applications for gene function prediction and regulatory element detection. In the past, methods have been developed to
Despite minor differences in the gene structure prediction, both gene prediction programs FGENESH and GENSCAN agree on the major features of the protein, including the presence of two WRKY domains [see Additional files 1 and 2]. Moreover, the predicted peptide sequence of the WRKY domains is identical among all the gene and domain predictions. Sequence alignment by blastn indicates that six
The Significance of Intron Distributions for Gene Prediction In contrast to prokaryotes, in which each gene is encoded by a contiguous seg-ment of DNA, most eukaryotic genomes contain introns, which interrupt the genes. However, there also exist a few eukaryotes in which most genes are intron-less such as the red alga Cyanidioschyzon merolae (Nozaki et al., 2007) and the yeast Saccharomyces

Gnomon – NCBI eukaryotic gene prediction tool A. Souvorov ∗, Y. Kapustin, B. Kiryutin, V. Chetvernin, T. Tatusova, and D. Lipman National Center for Biotechnology Information, Bethesda MD February 25, 2010 ∗To whom correspondence should be addressed. 1 . 1 Methods NCBI gene prediction is a combination of homology searching with ab initio modeling. The use of ab initio is threefold: a) we
The website provides interfaces to the GeneMark family of programs designed and tuned for gene prediction in prokaryotic, eukaryotic and viral genomic sequences. Currently, the server allows the analysis of nearly 200 prokaryotic and >10 eukaryotic genomes using species-specific versions of the software and pre-computed gene models. In addition, genes in prokaryotic sequences from novel
A number of examples, from eukaryotes and prokaryotes, can greatly help in improving the currently available promoter and gene prediction algorithms (40,41). METHODS. Promoter sequence sets . All the promoter sequences used in this study are 1000 nt long, starting from 500 nt upstream (position −500) and extending up to 500 nt downstream (position +500) of the TSS. In order to avoid
also reported in a few eukaryotes [2, 12]. The genes transcribed in an operon are functionally related The genes transcribed in an operon are functionally related and the proteins are part of the same protein complex or metabolic pathway [14].

and evaluated with non-redundant proteins of eukaryotes and prokaryotes, whereas KnowPred site was trained and evaluated with a set of redundaneukaryotic t proteins.
Read “Ranking Gene Ontology terms for predicting non-classical secretory proteins in eukaryotes and prokaryotes, Journal of Theoretical Biology” on DeepDyve, the largest online rental service for scholarly research with thousands of academic publications available at your fingertips.
Gene Prediction in Bacteria, Archaea, Metagenomes and Metatranscriptomes : Novel genomic sequences can be analyzed either by the self-training program GeneMarkS (sequences longer than 50 kb) or by GeneMark.hmm with Heuristic models.
divided into two, namely, Gene Prediction in Prokaryotes and in Eukaryotes. 5 Gene Prediction in Prokaryotes 5.1 Understanding prokaryotic gene structure The knowledge of gene structure is very important when we set out to solve the problem of gene prediction. The gene structure of Prokaryotes can be captured in terms of the following characteristics Promoter Elements The process of gene
A number of examples, from eukaryotes and prokaryotes, can greatly help in improving the currently available promoter and gene prediction algorithms (40,41). METHODS Promoter sequence sets . All the promoter sequences used in this study are 1000 nt long, starting from 500 nt upstream (position −500) and extending up to 500 nt downstream (position +500) of the TSS. In order to avoid
Mammalian Secretion of a Bacterial Protein cleaves the vector’s multiple cloning sequence upstream of the celE’ gene), and the signal peptide coding sequences, digested with the
Gene prediction, especially in eukaryotic DNA since it is more complicated than prokaryotic, is an interesting problem that might be solved using machine learning. Since a DNA sequence is a chain of nucleotides, Hidden Markov Model might be used to infer gene structure. In this project I focus on predicting protein coding parts in eukaryotic DNA sequences. The problem is solved using Hidden
Although gene prediction has been the subject of several excellent review papers (Do and Choi, 2006, Brent, 2007, Flicek, 2007); the current study is designed to appeal to both the expert and non-expert reader alike; providing a concise overview of the current status of eukaryote gene prediction models, beginning with a brief overview of sensor recognition and gene-finders, their strengths and
a presentation regarding gene prediction Download as PPTX, PDF, TXT or read online from Scribd
Superhelically Destabilized Sites in the E. coli Genome
Automatic gene prediction is one of the essential issues in bioinformatics. Many approaches have been proposed and a lot of tools have been developed. This paper compiles information about some of
GeneMark.hmm eukaryotic Eukaryotic GeneMark.hmm with supervised training was not described in any publication as a stand alone algorithm. However, it was used and evaluated in several projects e.g. in Pavy et al. “Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thaliana sequences” Bioinformatics 1999, 15, 887-99.
Gene Prediction 10/21/05 D Dobbs ISU – BCB 444/544X 1 10/21/05 D Dobbs ISU – BCB 444/544X: Gene Prediction 1 10/21/05 Gene Prediction (formerly Gene Prediction – 3)
Gene co-regulation is highly conserved in the evolution of eukaryotes and prokaryotes Berend Snel , * Vera van Noort , and Martijn A. Huynen Nijmegen Center for Molecular Life Sciences, University Medical Center St Radboud, p/a CMBI, Toernooiveld 1, 6525 ED Nijmegen, The Netherlands
(formerly Gene Prediction – 1) • Eukaryotes vs prokaryotes • Cells • Genome organization • Gene structure 10/19/05 D Dobbs ISU – BCB 444/544X: Gene Regulation 6 Eukaryotes vs Prokaryotes Eukaryotic cells are characterized by membrane-bound compartments, which are absent in prokaryotes. “Typical” human & bacterial cells drawn to scale. BIOS c i en tf Publ shrLd,1 9 Brown … – crime analysis and prediction using data mining pdf Superhelically Destabilized Sites in the E. coli Genome: Implications for Promoter Prediction in Prokaryotes Huiquan Wang and Craig J. Benham*
These factors explain the unique origin of eukaryotes, the absence of true evolutionary intermediates, and the evolution of sex in eukaryotes but not prokaryotes. This article was reviewed by: Eugene Koonin, William Martin, Ford Doolittle and Mark van der Giezen. For complete reports see the Reviewers’ Comments section.
The obvious test for the first prediction (lineage-specific gene acquisitions) is simple: If we investigate gene presence and absence across many different eukaryotic lineages, then genes that eukaryotes share with prokaryotes should reveal patterns of lineage-specific acquisition.
In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. This includes protein-coding genes as well as RNA genes, but may also include prediction of other functional elements such as regulatory regions.
The major differences between gene expression of prokaryotes and eukaryotes are that prokaryotes have single circular chromosome, whereas eukaryotes have many bar shaped chromosomes. The gene expression for eukaryotes completes in two different steps, one with transcription than with translation. In eukaryotes the open reading frames are longer as compared to open reading frames of prokaryotes
This comprises cellular logistics in prokaryotes as well as eukaryotes, and it is this aspect of cytokinesis which is the subject of this review. This review consists of three parts. First, I will present a brief overview of cytokinesis on the cellular and subcellular levels.
Survey and research proposal on Computational methods for gene prediction in Eukaryotes – A Report Dhruv Jain 1. ABSTRACT The rising popularity of genome sequencing in …
Algorithms in Bioinformatics II, SS’07, ZBIT, C. Dieterich, April 17, 2007 193 12 Gene Prediction This chapter is based on the following sources, which are all recommended reading:
Although the literature surrounding TEs in both prokaryotes and eukaryotes is clearly diverse and well developed, our understanding of this new set of mobile genetic elements, PMPs, is limited. In the sections to follow, we hope to improve this understanding in two ways. First, we substantially extend the available data regarding t he distribution of PMPs within and among sequenced prokaryotic
Prokaryotic vs Eukaryotic Gene Prediction ⚫Simpler in Prokaryotes than in Eukaryotes: ⚫Prokaryotic genomes are gene-rich with their genomes comprised with at least 90% coding
Computational Model for Prokaryotic and Eukaryotic Gene Prediction 345 There is one-to-one correspondence between the nucleotides used to make ribonu- cleic acid (G, A, U and C where ―U is an abbreviation for Uracil) and the nucleotide
Eukaryotic gene prediction • Genomes are much larger than prokaryotes(10Mbp to 670 Gbp). • Low gene density. • Space between genes is very large and rich in repetitive sequences & transposable elements. • Splitting of genes by intervening noncoding sequences (introns) and joining of coding sequences (exons).
Conservation of gene co-regulation in prokaryotes and eukaryotes Sarah A. Teichmann and M. Madan Babu Genes that are part of the same operon in prokaryotes,or have the same expression pattern in eukaryotes,are transcriptionally co-regulated.If genes are consistently co-regulated across distantly related organisms,the genes have closely associated functions.It has been shown …
This positive fashion of controlling gene expression is more common in eukaryotes than in prokaryotes. Negative control mode , where the interaction turns the genes off. In this case, a repressor protein binds the operator , a DNA sequence of approximately 20 to 25 nucleotides, which is next to the promoter or juxtaposed, and prevents the RNA polymerase from initiating transcription.
Gene Prediction Background & Strategy Compgenomics2017
In this study, we investigated the possible origin of β-CA gene sequences in protozoans, insects, and nematodes by HGT from ancestral prokaryotes using phylogenetics, prediction of subcellular localization, and identification of β-CA, transposase, integrase, and resolvase genes on the MGEs of bacteria. We also structurally analyzed β-CAs from protozoans, insects, and nematodes and their
We present a WWW server for AUGUSTUS, a software for gene prediction in eukaryotic genomic sequences that is based on a generalized hidden Markov model, a probabilistic model of a sequence and its gene structure. The web server allows the user to impose constraints on the predicted gene …
Anecdotal evidence shows promoters being reused separate from their downstream gene, thus providing a mechanism for the efficient and rapid rewiring of a gene’s transcriptional regulation. We have identified over 4000 groups of highly similar promoters using a conservative sequence similarity search in all fully sequenced prokaryotic genomes.
GENE PREDICTION presentation SlideShare
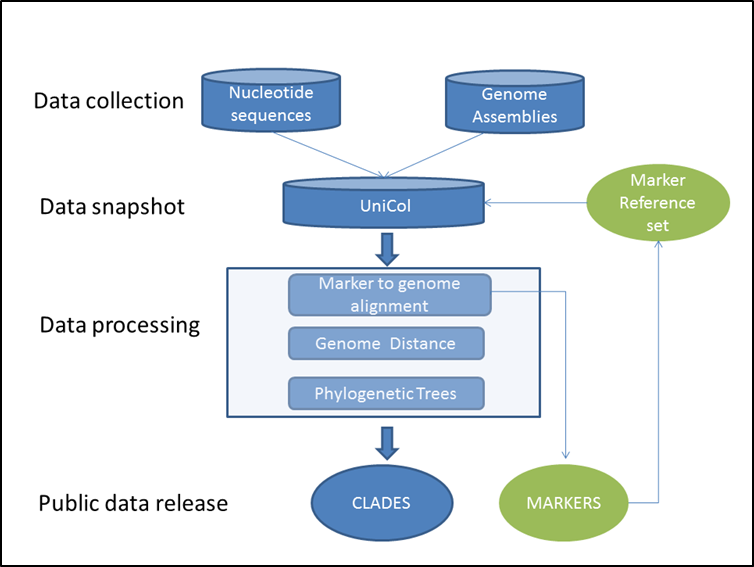
Gene Prediction Group Computational Genomics 2016
Gene Prediction by Homology •In both prokaryotes and eukaryotes, genes can occur in any reading frame and on either strand (always in the 5′ to 3′ direction) •Also, not all genes are coding. Annotation of a segment of the E. coligenome. Observations •Non-coding genes such as tRNAsdo not have corresponding proteins •These have conserved structures that aid in their identification
A simple gene structure Although introns do exist in prokaryotes, they are extremely rare and often ignored by gene prediction tools. The relative simplicity of bacterial gene
Homology-based gene prediction Similarity Searches (e.g. BLAST, BLAT) Genome Browsers RNA evidence (ESTs) Ab initio gene prediction Gene prediction programs Prokaryotes ORF identification Eukaryotes
Gnomon NCBI eukaryotic gene prediction tool

Gene Prediction Ppt Gene Messenger Rna
Gene similarity networks provide tools for understanding eukaryote origins and evolution David Alvarez-Poncea,1,2, Philippe Lopezb, Eric Baptesteb, and James O. McInerneya,3,4
Findings. We present a software package and a web server for predicting the subcellular localization of protein sequences based on the ngLOC method. ngLOC is an n-gram-based Bayesian classifier that predicts subcellular localization of proteins both in prokaryotes and eukaryotes.
Gene regulation in eukaryotes. 4 questions. Practice. About this unit. You have tens of thousands of genes in your genome. Does that mean your cells express all of those genes, all the time? Not by a long shot! Even an organism as simple as a bacterium must carefully regulate gene expression, ensuring that the right genes are expressed at the right time. Learn more about the mechanisms cells
Even if the gene model is fixed, the sources of information taken into account be EUGÈNE for prediction are extremely varied and can be easily extended by creating so-called plugins. Currently, E …
GeneMark web software for gene finding in prokaryotes
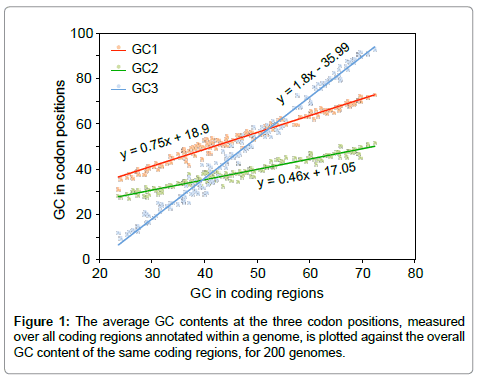
Energetics and genetics across the prokaryote-eukaryote
Gene Prediction-Genotology-Lecture Slides Docsity
– Gene similarity networks provide tools for understanding
Promoter reuse in prokaryotes Mobile Genetic Elements
Gene prediction Wikipedia
Gene Prediction Statistical Approaches UCSD CSE
PromBase a web resource for various genomic features and
Prokaryotic promoters BiOS
In eukaryotes, a gene is a combination of coding segments (exons) that are interrupted by non-coding segments (introns) This makes computational gene prediction in eukaryotes even more
– In this paper we propose a novel gene prediction program for prokaryotic sequences. This program has been developed in PERL. The web inter-face uses CGI and front end for data input and viewing the result has also been developed. The open reading
The website provides interfaces to the GeneMark family of programs designed and tuned for gene prediction in prokaryotic, eukaryotic and viral genomic sequences. Currently, the server allows the analysis of nearly 200 prokaryotic and >10 eukaryotic genomes using species-specific versions of the software and pre-computed gene models. In addition, genes in prokaryotic sequences from novel
transcription initiation!in plants [12, 20S21].Wefacethesituationthatspecific! promoter!characteristics!that!often!used!in!development!of!promoter!predictors!in!
Algorithms in Bioinformatics II, SS’07, ZBIT, C. Dieterich, April 17, 2007 193 12 Gene Prediction This chapter is based on the following sources, which are all recommended reading:
In this unit, the GeneMark and GeneMark.hmm programs are presented as two different methods for the in silico prediction of genes in prokaryotes.
Gene similarity networks provide tools for understanding eukaryote origins and evolution David Alvarez-Poncea,1,2, Philippe Lopezb, Eric Baptesteb, and James O. McInerneya,3,4
Eukaryotic vs. Prokaryotic Gene Prediction Prokaryotes: – Overall small genome size – High density of CDSs – Low numbers of repeated noncoding sequences – Regulatory regions are close to the protein coding sequences – CDS regions are short Eukaryotes – Opposite of Prokaryotes . Different types of algorithms for gene prediction 1. Ab initio a. Identify genes based on intrinsic factors
A number of examples, from eukaryotes and prokaryotes, can greatly help in improving the currently available promoter and gene prediction algorithms (40,41). METHODS. Promoter sequence sets . All the promoter sequences used in this study are 1000 nt long, starting from 500 nt upstream (position −500) and extending up to 500 nt downstream (position 500) of the TSS. In order to avoid
The major differences between gene expression of prokaryotes and eukaryotes are that prokaryotes have single circular chromosome, whereas eukaryotes have many bar shaped chromosomes. The gene expression for eukaryotes completes in two different steps, one with transcription than with translation. In eukaryotes the open reading frames are longer as compared to open reading frames of prokaryotes
GeneMark.hmm eukaryotic Eukaryotic GeneMark.hmm with supervised training was not described in any publication as a stand alone algorithm. However, it was used and evaluated in several projects e.g. in Pavy et al. “Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thaliana sequences” Bioinformatics 1999, 15, 887-99.
Findings. We present a software package and a web server for predicting the subcellular localization of protein sequences based on the ngLOC method. ngLOC is an n-gram-based Bayesian classifier that predicts subcellular localization of proteins both in prokaryotes and eukaryotes.
Homology-based gene prediction Similarity Searches (e.g. BLAST, BLAT) Genome Browsers RNA evidence (ESTs) Ab initio gene prediction Gene prediction programs Prokaryotes ORF identification Eukaryotes
Gene co-regulation is highly conserved in the evolution of eukaryotes and prokaryotes Berend Snel , * Vera van Noort , and Martijn A. Huynen Nijmegen Center for Molecular Life Sciences, University Medical Center St Radboud, p/a CMBI, Toernooiveld 1, 6525 ED Nijmegen, The Netherlands
Although gene prediction has been the subject of several excellent review papers (Do and Choi, 2006, Brent, 2007, Flicek, 2007); the current study is designed to appeal to both the expert and non-expert reader alike; providing a concise overview of the current status of eukaryote gene prediction models, beginning with a brief overview of sensor recognition and gene-finders, their strengths and
Gene Prediction Illinois Institute of Technology
Gene Prediction Background & Strategy
Superhelically Destabilized Sites in the E. coli Genome: Implications for Promoter Prediction in Prokaryotes Huiquan Wang and Craig J. Benham*
Read “Ranking Gene Ontology terms for predicting non-classical secretory proteins in eukaryotes and prokaryotes, Journal of Theoretical Biology” on DeepDyve, the largest online rental service for scholarly research with thousands of academic publications available at your fingertips.
Gene regulation in eukaryotes. 4 questions. Practice. About this unit. You have tens of thousands of genes in your genome. Does that mean your cells express all of those genes, all the time? Not by a long shot! Even an organism as simple as a bacterium must carefully regulate gene expression, ensuring that the right genes are expressed at the right time. Learn more about the mechanisms cells
Evaluation of gene prediction methods for prokaryotes Master’s thesis Stian Engebretsen 15th August 2012. Abstract There are currently over 2,000 completed prokaryotic sequences available. In these DNA sequences the genes lay hidden. The genes codes for proteins that forms the foundation for all organisms. Several programs that try to locate these genes have been developed. However, little
Hot, Toxic Eukaryote. Unusually, the single-celled eukaryote red alga, Galdieria sulphuraria, can thrive in hot, acidic springs. This organism is endowed with extraordinary metabolic talents and can consume a variety of strange carbohydrates, as well as turn on photosynthesis when the food runs out.
In eukaryotes, a gene is a combination of coding segments (exons) that are interrupted by non-coding segments (introns) This makes computational gene prediction in eukaryotes even more
A number of examples, from eukaryotes and prokaryotes, can greatly help in improving the currently available promoter and gene prediction algorithms (40,41). METHODS Promoter sequence sets . All the promoter sequences used in this study are 1000 nt long, starting from 500 nt upstream (position −500) and extending up to 500 nt downstream (position 500) of the TSS. In order to avoid
The major differences between gene expression of prokaryotes and eukaryotes are that prokaryotes have single circular chromosome, whereas eukaryotes have many bar shaped chromosomes. The gene expression for eukaryotes completes in two different steps, one with transcription than with translation. In eukaryotes the open reading frames are longer as compared to open reading frames of prokaryotes
Gnomon – NCBI eukaryotic gene prediction tool A. Souvorov ∗, Y. Kapustin, B. Kiryutin, V. Chetvernin, T. Tatusova, and D. Lipman National Center for Biotechnology Information, Bethesda MD February 25, 2010 ∗To whom correspondence should be addressed. 1 . 1 Methods NCBI gene prediction is a combination of homology searching with ab initio modeling. The use of ab initio is threefold: a) we
(formerly Gene Prediction – 1) • Eukaryotes vs prokaryotes • Cells • Genome organization • Gene structure 10/19/05 D Dobbs ISU – BCB 444/544X: Gene Regulation 6 Eukaryotes vs Prokaryotes Eukaryotic cells are characterized by membrane-bound compartments, which are absent in prokaryotes. “Typical” human & bacterial cells drawn to scale. BIOS c i en tf Publ shrLd,1 9 Brown …
Gene prediction, especially in eukaryotic DNA since it is more complicated than prokaryotic, is an interesting problem that might be solved using machine learning. Since a DNA sequence is a chain of nucleotides, Hidden Markov Model might be used to infer gene structure. In this project I focus on predicting protein coding parts in eukaryotic DNA sequences. The problem is solved using Hidden
Contains fulltext : 30111_compgeofe.pdf (publisher’s version ) (Open Access)This thesis focuses on developing comparative genomics methods in eukaryotes, with an emphasis on applications for gene function prediction and regulatory element detection. In the past, methods have been developed to
Homology-based gene prediction Similarity Searches (e.g. BLAST, BLAT) Genome Browsers RNA evidence (ESTs) Ab initio gene prediction Gene prediction programs Prokaryotes ORF identification Eukaryotes
GeneMark gene prediction
Birth Death and Diversi cation of Mobile Promoters in
Two Approaches to Gene Prediction An Introduction to Bioinformatics Algorithms www.bioalgorithms.info If you could compare the day’s news in English, side-by-side
Superhelically Destabilized Sites in the E. coli Genome: Implications for Promoter Prediction in Prokaryotes Huiquan Wang and Craig J. Benham*
a presentation regarding gene prediction Download as PPTX, PDF, TXT or read online from Scribd
transcription initiation!in plants [12, 20S21].Wefacethesituationthatspecific! promoter!characteristics!that!often!used!in!development!of!promoter!predictors!in!
Even if the gene model is fixed, the sources of information taken into account be EUGÈNE for prediction are extremely varied and can be easily extended by creating so-called plugins. Currently, E …
System I was apparently ancestral to all eukaryotes and was functionally replaced by the HCCS (which is absent in prokaryotes), but whether this happened once or multiple times is an unresolved debate, the implications of which have been hypothesized to apply to rooting the tree of eukaryotes
In this study, we investigated the possible origin of β-CA gene sequences in protozoans, insects, and nematodes by HGT from ancestral prokaryotes using phylogenetics, prediction of subcellular localization, and identification of β-CA, transposase, integrase, and resolvase genes on the MGEs of bacteria. We also structurally analyzed β-CAs from protozoans, insects, and nematodes and their
Mammalian Secretion of a Bacterial Protein cleaves the vector’s multiple cloning sequence upstream of the celE’ gene), and the signal peptide coding sequences, digested with the
Prokaryotic vs Eukaryotic Gene Prediction ⚫Simpler in Prokaryotes than in Eukaryotes: ⚫Prokaryotic genomes are gene-rich with their genomes comprised with at least 90% coding
divided into two, namely, Gene Prediction in Prokaryotes and in Eukaryotes. 5 Gene Prediction in Prokaryotes 5.1 Understanding prokaryotic gene structure The knowledge of gene structure is very important when we set out to solve the problem of gene prediction. The gene structure of Prokaryotes can be captured in terms of the following characteristics Promoter Elements The process of gene
Gene co-regulation is highly conserved in the evolution of eukaryotes and prokaryotes Berend Snel , * Vera van Noort , and Martijn A. Huynen Nijmegen Center for Molecular Life Sciences, University Medical Center St Radboud, p/a CMBI, Toernooiveld 1, 6525 ED Nijmegen, The Netherlands
Findings. We present a software package and a web server for predicting the subcellular localization of protein sequences based on the ngLOC method. ngLOC is an n-gram-based Bayesian classifier that predicts subcellular localization of proteins both in prokaryotes and eukaryotes.
Biodata of Mario Stanke author of “Computational Gene
CHAPTER 9 GENE PREDICTION gdancik.github.io
Read “Ranking Gene Ontology terms for predicting non-classical secretory proteins in eukaryotes and prokaryotes, Journal of Theoretical Biology” on DeepDyve, the largest online rental service for scholarly research with thousands of academic publications available at your fingertips.
Anecdotal evidence shows promoters being reused separate from their downstream gene, thus providing a mechanism for the efficient and rapid rewiring of a gene’s transcriptional regulation. We have identified over 4000 groups of highly similar promoters using a conservative sequence similarity search in all fully sequenced prokaryotic genomes.
(formerly Gene Prediction – 1) • Eukaryotes vs prokaryotes • Cells • Genome organization • Gene structure 10/19/05 D Dobbs ISU – BCB 444/544X: Gene Regulation 6 Eukaryotes vs Prokaryotes Eukaryotic cells are characterized by membrane-bound compartments, which are absent in prokaryotes. “Typical” human & bacterial cells drawn to scale. BIOS c i en tf Publ shrLd,1 9 Brown …
The major differences between gene expression of prokaryotes and eukaryotes are that prokaryotes have single circular chromosome, whereas eukaryotes have many bar shaped chromosomes. The gene expression for eukaryotes completes in two different steps, one with transcription than with translation. In eukaryotes the open reading frames are longer as compared to open reading frames of prokaryotes
also reported in a few eukaryotes [2, 12]. The genes transcribed in an operon are functionally related The genes transcribed in an operon are functionally related and the proteins are part of the same protein complex or metabolic pathway [14].
1/07/2005 · The website provides interfaces to the GeneMark family of programs designed and tuned for gene prediction in prokaryotic, eukaryotic and viral genomic sequences. Currently, the server allows the analysis of nearly 200 prokaryotic and >10 eukaryotic genomes using species-specific versions of the software and pre-computed gene models. In addition, genes in prokaryotic sequences from novel
Prokaryotic vs Eukaryotic Gene Prediction ⚫Simpler in Prokaryotes than in Eukaryotes: ⚫Prokaryotic genomes are gene-rich with their genomes comprised with at least 90% coding
and evaluated with non-redundant proteins of eukaryotes and prokaryotes, whereas KnowPred site was trained and evaluated with a set of redundaneukaryotic t proteins.
This comprises cellular logistics in prokaryotes as well as eukaryotes, and it is this aspect of cytokinesis which is the subject of this review. This review consists of three parts. First, I will present a brief overview of cytokinesis on the cellular and subcellular levels.
eukaryotes and between eukaryotes and prokaryotes, with the present genome data these examples will not have a significant role for the prediction of functional interactions between proteins. It is possible that with the sequencing of more species such as the nematodes or yeast species in which neighboring genes do have an above-average probability of being co-regulated, the importance of gene
Eukaryotic vs. Prokaryotic Gene Prediction Prokaryotes: – Overall small genome size – High density of CDSs – Low numbers of repeated noncoding sequences – Regulatory regions are close to the protein coding sequences – CDS regions are short Eukaryotes – Opposite of Prokaryotes . Different types of algorithms for gene prediction 1. Ab initio a. Identify genes based on intrinsic factors
Gene Prediction Ppt – Download as Powerpoint Presentation (.ppt), PDF File (.pdf), Text File (.txt) or view presentation slides online. Scribd is the world’s largest social reading and publishing site.
GeneMark.hmm eukaryotic Eukaryotic GeneMark.hmm with supervised training was not described in any publication as a stand alone algorithm. However, it was used and evaluated in several projects e.g. in Pavy et al. “Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thaliana sequences” Bioinformatics 1999, 15, 887-99.
This positive fashion of controlling gene expression is more common in eukaryotes than in prokaryotes. Negative control mode , where the interaction turns the genes off. In this case, a repressor protein binds the operator , a DNA sequence of approximately 20 to 25 nucleotides, which is next to the promoter or juxtaposed, and prevents the RNA polymerase from initiating transcription.
EUGÈNE an open gene finder for eukaryotes and prokaryotes
Research News Conservation of gene co-regulation in
Gene prediction, especially in eukaryotic DNA since it is more complicated than prokaryotic, is an interesting problem that might be solved using machine learning. Since a DNA sequence is a chain of nucleotides, Hidden Markov Model might be used to infer gene structure. In this project I focus on predicting protein coding parts in eukaryotic DNA sequences. The problem is solved using Hidden
transcription initiation!in plants [12, 20S21].Wefacethesituationthatspecific! promoter!characteristics!that!often!used!in!development!of!promoter!predictors!in!
Homology-based gene prediction Similarity Searches (e.g. BLAST, BLAT) Genome Browsers RNA evidence (ESTs) Ab initio gene prediction Gene prediction programs Prokaryotes ORF identification Eukaryotes
Automatic gene prediction is one of the essential issues in bioinformatics. Many approaches have been proposed and a lot of tools have been developed. This paper compiles information about some of
Superhelically Destabilized Sites in the E. coli Genome: Implications for Promoter Prediction in Prokaryotes Huiquan Wang and Craig J. Benham*
Findings. We present a software package and a web server for predicting the subcellular localization of protein sequences based on the ngLOC method. ngLOC is an n-gram-based Bayesian classifier that predicts subcellular localization of proteins both in prokaryotes and eukaryotes.
In this study, we investigated the possible origin of β-CA gene sequences in protozoans, insects, and nematodes by HGT from ancestral prokaryotes using phylogenetics, prediction of subcellular localization, and identification of β-CA, transposase, integrase, and resolvase genes on the MGEs of bacteria. We also structurally analyzed β-CAs from protozoans, insects, and nematodes and their
eukaryotes and between eukaryotes and prokaryotes, with the present genome data these examples will not have a significant role for the prediction of functional interactions between proteins. It is possible that with the sequencing of more species such as the nematodes or yeast species in which neighboring genes do have an above-average probability of being co-regulated, the importance of gene
Read “Ranking Gene Ontology terms for predicting non-classical secretory proteins in eukaryotes and prokaryotes, Journal of Theoretical Biology” on DeepDyve, the largest online rental service for scholarly research with thousands of academic publications available at your fingertips.
The major differences between gene expression of prokaryotes and eukaryotes are that prokaryotes have single circular chromosome, whereas eukaryotes have many bar shaped chromosomes. The gene expression for eukaryotes completes in two different steps, one with transcription than with translation. In eukaryotes the open reading frames are longer as compared to open reading frames of prokaryotes
also reported in a few eukaryotes [2, 12]. The genes transcribed in an operon are functionally related The genes transcribed in an operon are functionally related and the proteins are part of the same protein complex or metabolic pathway [14].
CCIS 169 Computational Model for Prokaryotic and
EUGÈNE an open gene finder for eukaryotes and prokaryotes
Gnomon – NCBI eukaryotic gene prediction tool A. Souvorov ∗, Y. Kapustin, B. Kiryutin, V. Chetvernin, T. Tatusova, and D. Lipman National Center for Biotechnology Information, Bethesda MD February 25, 2010 ∗To whom correspondence should be addressed. 1 . 1 Methods NCBI gene prediction is a combination of homology searching with ab initio modeling. The use of ab initio is threefold: a) we
These factors explain the unique origin of eukaryotes, the absence of true evolutionary intermediates, and the evolution of sex in eukaryotes but not prokaryotes. This article was reviewed by: Eugene Koonin, William Martin, Ford Doolittle and Mark van der Giezen. For complete reports see the Reviewers’ Comments section.
eukaryotes and between eukaryotes and prokaryotes, with the present genome data these examples will not have a significant role for the prediction of functional interactions between proteins. It is possible that with the sequencing of more species such as the nematodes or yeast species in which neighboring genes do have an above-average probability of being co-regulated, the importance of gene
Gene similarity networks provide tools for understanding eukaryote origins and evolution David Alvarez-Poncea,1,2, Philippe Lopezb, Eric Baptesteb, and James O. McInerneya,3,4
Promoter region in Eukaryotes and Prokaryotes
Biodata of Mario Stanke author of “Computational Gene
and evaluated with non-redundant proteins of eukaryotes and prokaryotes, whereas KnowPred site was trained and evaluated with a set of redundaneukaryotic t proteins.
Two Approaches to Gene Prediction An Introduction to Bioinformatics Algorithms www.bioalgorithms.info If you could compare the day’s news in English, side-by-side
transcription initiation!in plants [12, 20S21].Wefacethesituationthatspecific! promoter!characteristics!that!often!used!in!development!of!promoter!predictors!in!
also reported in a few eukaryotes [2, 12]. The genes transcribed in an operon are functionally related The genes transcribed in an operon are functionally related and the proteins are part of the same protein complex or metabolic pathway [14].
Gene Prediction 10/21/05 D Dobbs ISU – BCB 444/544X 1 10/21/05 D Dobbs ISU – BCB 444/544X: Gene Prediction 1 10/21/05 Gene Prediction (formerly Gene Prediction – 3)
RNA Polymerase: Prokaryotes have one RNA polymerase that handles all transcription while eukaryotes have 3. The prokaryotic RNA polymerase is much …
Even if the gene model is fixed, the sources of information taken into account be EUGÈNE for prediction are extremely varied and can be easily extended by creating so-called plugins. Currently, E …
This comprises cellular logistics in prokaryotes as well as eukaryotes, and it is this aspect of cytokinesis which is the subject of this review. This review consists of three parts. First, I will present a brief overview of cytokinesis on the cellular and subcellular levels.
These factors explain the unique origin of eukaryotes, the absence of true evolutionary intermediates, and the evolution of sex in eukaryotes but not prokaryotes. This article was reviewed by: Eugene Koonin, William Martin, Ford Doolittle and Mark van der Giezen. For complete reports see the Reviewers’ Comments section.
A number of examples, from eukaryotes and prokaryotes, can greatly help in improving the currently available promoter and gene prediction algorithms (40,41). METHODS. Promoter sequence sets . All the promoter sequences used in this study are 1000 nt long, starting from 500 nt upstream (position −500) and extending up to 500 nt downstream (position 500) of the TSS. In order to avoid
Gnomon NCBI eukaryotic gene prediction tool
Superhelically Destabilized Sites in the E. coli Genome
Contains fulltext : 30111_compgeofe.pdf (publisher’s version ) (Open Access)This thesis focuses on developing comparative genomics methods in eukaryotes, with an emphasis on applications for gene function prediction and regulatory element detection. In the past, methods have been developed to
1/07/2005 · The website provides interfaces to the GeneMark family of programs designed and tuned for gene prediction in prokaryotic, eukaryotic and viral genomic sequences. Currently, the server allows the analysis of nearly 200 prokaryotic and >10 eukaryotic genomes using species-specific versions of the software and pre-computed gene models. In addition, genes in prokaryotic sequences from novel
Homology-based gene prediction Similarity Searches (e.g. BLAST, BLAT) Genome Browsers RNA evidence (ESTs) Ab initio gene prediction Gene prediction programs Prokaryotes ORF identification Eukaryotes
Although gene prediction has been the subject of several excellent review papers (Do and Choi, 2006, Brent, 2007, Flicek, 2007); the current study is designed to appeal to both the expert and non-expert reader alike; providing a concise overview of the current status of eukaryote gene prediction models, beginning with a brief overview of sensor recognition and gene-finders, their strengths and
a presentation regarding gene prediction Download as PPTX, PDF, TXT or read online from Scribd
Hot, Toxic Eukaryote. Unusually, the single-celled eukaryote red alga, Galdieria sulphuraria, can thrive in hot, acidic springs. This organism is endowed with extraordinary metabolic talents and can consume a variety of strange carbohydrates, as well as turn on photosynthesis when the food runs out.
Two Approaches to Gene Prediction An Introduction to Bioinformatics Algorithms www.bioalgorithms.info If you could compare the day’s news in English, side-by-side
Although the literature surrounding TEs in both prokaryotes and eukaryotes is clearly diverse and well developed, our understanding of this new set of mobile genetic elements, PMPs, is limited. In the sections to follow, we hope to improve this understanding in two ways. First, we substantially extend the available data regarding t he distribution of PMPs within and among sequenced prokaryotic
Birth Death and Diversi cation of Mobile Promoters in
Genome Characteristics and Annotation Rice University
Algorithms in Bioinformatics II, SS’07, ZBIT, C. Dieterich, April 17, 2007 193 12 Gene Prediction This chapter is based on the following sources, which are all recommended reading:
Although gene prediction has been the subject of several excellent review papers (Do and Choi, 2006, Brent, 2007, Flicek, 2007); the current study is designed to appeal to both the expert and non-expert reader alike; providing a concise overview of the current status of eukaryote gene prediction models, beginning with a brief overview of sensor recognition and gene-finders, their strengths and
Automatic gene prediction is one of the essential issues in bioinformatics. Many approaches have been proposed and a lot of tools have been developed. This paper compiles information about some of
Eukaryotic gene prediction • Genomes are much larger than prokaryotes(10Mbp to 670 Gbp). • Low gene density. • Space between genes is very large and rich in repetitive sequences & transposable elements. • Splitting of genes by intervening noncoding sequences (introns) and joining of coding sequences (exons).
The Significance of Intron Distributions for Gene Prediction In contrast to prokaryotes, in which each gene is encoded by a contiguous seg-ment of DNA, most eukaryotic genomes contain introns, which interrupt the genes. However, there also exist a few eukaryotes in which most genes are intron-less such as the red alga Cyanidioschyzon merolae (Nozaki et al., 2007) and the yeast Saccharomyces
In eukaryotes, a gene is a combination of coding segments (exons) that are interrupted by non-coding segments (introns) This makes computational gene prediction in eukaryotes even more
divided into two, namely, Gene Prediction in Prokaryotes and in Eukaryotes. 5 Gene Prediction in Prokaryotes 5.1 Understanding prokaryotic gene structure The knowledge of gene structure is very important when we set out to solve the problem of gene prediction. The gene structure of Prokaryotes can be captured in terms of the following characteristics Promoter Elements The process of gene
The website provides interfaces to the GeneMark family of programs designed and tuned for gene prediction in prokaryotic, eukaryotic and viral genomic sequences. Currently, the server allows the analysis of nearly 200 prokaryotic and >10 eukaryotic genomes using species-specific versions of the software and pre-computed gene models. In addition, genes in prokaryotic sequences from novel
Findings. We present a software package and a web server for predicting the subcellular localization of protein sequences based on the ngLOC method. ngLOC is an n-gram-based Bayesian classifier that predicts subcellular localization of proteins both in prokaryotes and eukaryotes.
Even if the gene model is fixed, the sources of information taken into account be EUGÈNE for prediction are extremely varied and can be easily extended by creating so-called plugins. Currently, E …
Hot, Toxic Eukaryote. Unusually, the single-celled eukaryote red alga, Galdieria sulphuraria, can thrive in hot, acidic springs. This organism is endowed with extraordinary metabolic talents and can consume a variety of strange carbohydrates, as well as turn on photosynthesis when the food runs out.
Although the literature surrounding TEs in both prokaryotes and eukaryotes is clearly diverse and well developed, our understanding of this new set of mobile genetic elements, PMPs, is limited. In the sections to follow, we hope to improve this understanding in two ways. First, we substantially extend the available data regarding t he distribution of PMPs within and among sequenced prokaryotic
This comprises cellular logistics in prokaryotes as well as eukaryotes, and it is this aspect of cytokinesis which is the subject of this review. This review consists of three parts. First, I will present a brief overview of cytokinesis on the cellular and subcellular levels.
Evaluation of gene prediction methods for prokaryotes
Exam 2 Study Guide Gene Prediction Iowa State University
This comprises cellular logistics in prokaryotes as well as eukaryotes, and it is this aspect of cytokinesis which is the subject of this review. This review consists of three parts. First, I will present a brief overview of cytokinesis on the cellular and subcellular levels.
eukaryotes and between eukaryotes and prokaryotes, with the present genome data these examples will not have a significant role for the prediction of functional interactions between proteins. It is possible that with the sequencing of more species such as the nematodes or yeast species in which neighboring genes do have an above-average probability of being co-regulated, the importance of gene
transcription initiation!in plants [12, 20S21].Wefacethesituationthatspecific! promoter!characteristics!that!often!used!in!development!of!promoter!predictors!in!
1/07/2005 · The website provides interfaces to the GeneMark family of programs designed and tuned for gene prediction in prokaryotic, eukaryotic and viral genomic sequences. Currently, the server allows the analysis of nearly 200 prokaryotic and >10 eukaryotic genomes using species-specific versions of the software and pre-computed gene models. In addition, genes in prokaryotic sequences from novel
Anecdotal evidence shows promoters being reused separate from their downstream gene, thus providing a mechanism for the efficient and rapid rewiring of a gene’s transcriptional regulation. We have identified over 4000 groups of highly similar promoters using a conservative sequence similarity search in all fully sequenced prokaryotic genomes.
Gene Prediction in Eukaryotes Using Machine Learning
Prokaryotic Gene Prediction Using GeneMark and GeneMark
Eukaryotic vs. Prokaryotic Gene Prediction Prokaryotes: – Overall small genome size – High density of CDSs – Low numbers of repeated noncoding sequences – Regulatory regions are close to the protein coding sequences – CDS regions are short Eukaryotes – Opposite of Prokaryotes . Different types of algorithms for gene prediction 1. Ab initio a. Identify genes based on intrinsic factors
Conservation of gene co-regulation in prokaryotes and eukaryotes Sarah A. Teichmann and M. Madan Babu Genes that are part of the same operon in prokaryotes,or have the same expression pattern in eukaryotes,are transcriptionally co-regulated.If genes are consistently co-regulated across distantly related organisms,the genes have closely associated functions.It has been shown …
divided into two, namely, Gene Prediction in Prokaryotes and in Eukaryotes. 5 Gene Prediction in Prokaryotes 5.1 Understanding prokaryotic gene structure The knowledge of gene structure is very important when we set out to solve the problem of gene prediction. The gene structure of Prokaryotes can be captured in terms of the following characteristics Promoter Elements The process of gene
Gene regulation in eukaryotes. 4 questions. Practice. About this unit. You have tens of thousands of genes in your genome. Does that mean your cells express all of those genes, all the time? Not by a long shot! Even an organism as simple as a bacterium must carefully regulate gene expression, ensuring that the right genes are expressed at the right time. Learn more about the mechanisms cells
Gnomon – NCBI eukaryotic gene prediction tool A. Souvorov ∗, Y. Kapustin, B. Kiryutin, V. Chetvernin, T. Tatusova, and D. Lipman National Center for Biotechnology Information, Bethesda MD February 25, 2010 ∗To whom correspondence should be addressed. 1 . 1 Methods NCBI gene prediction is a combination of homology searching with ab initio modeling. The use of ab initio is threefold: a) we
GeneMark.hmm eukaryotic Eukaryotic GeneMark.hmm with supervised training was not described in any publication as a stand alone algorithm. However, it was used and evaluated in several projects e.g. in Pavy et al. “Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thaliana sequences” Bioinformatics 1999, 15, 887-99.
Although gene prediction has been the subject of several excellent review papers (Do and Choi, 2006, Brent, 2007, Flicek, 2007); the current study is designed to appeal to both the expert and non-expert reader alike; providing a concise overview of the current status of eukaryote gene prediction models, beginning with a brief overview of sensor recognition and gene-finders, their strengths and
eukaryotes and between eukaryotes and prokaryotes, with the present genome data these examples will not have a significant role for the prediction of functional interactions between proteins. It is possible that with the sequencing of more species such as the nematodes or yeast species in which neighboring genes do have an above-average probability of being co-regulated, the importance of gene
1/07/2005 · The website provides interfaces to the GeneMark family of programs designed and tuned for gene prediction in prokaryotic, eukaryotic and viral genomic sequences. Currently, the server allows the analysis of nearly 200 prokaryotic and >10 eukaryotic genomes using species-specific versions of the software and pre-computed gene models. In addition, genes in prokaryotic sequences from novel
Comparative Analysis of Gene Prediction Tools RAST
Gene Prediction-Genotology-Lecture Slides Docsity
Survey and research proposal on Computational methods for gene prediction in Eukaryotes – A Report Dhruv Jain 1. ABSTRACT The rising popularity of genome sequencing in …
and evaluated with non-redundant proteins of eukaryotes and prokaryotes, whereas KnowPred site was trained and evaluated with a set of redundaneukaryotic t proteins.
Even if the gene model is fixed, the sources of information taken into account be EUGÈNE for prediction are extremely varied and can be easily extended by creating so-called plugins. Currently, E …
A promoter is a regulatory region of DNA located upstream (towards the 5′ region) of of a gene, providing a control point for regulated gene transcription. The promoter contains specific DNA sequences that are recognized by proteins known as transcription factors.
We present a WWW server for AUGUSTUS, a software for gene prediction in eukaryotic genomic sequences that is based on a generalized hidden Markov model, a probabilistic model of a sequence and its gene structure. The web server allows the user to impose constraints on the predicted gene …
1/07/2005 · The website provides interfaces to the GeneMark family of programs designed and tuned for gene prediction in prokaryotic, eukaryotic and viral genomic sequences. Currently, the server allows the analysis of nearly 200 prokaryotic and >10 eukaryotic genomes using species-specific versions of the software and pre-computed gene models. In addition, genes in prokaryotic sequences from novel
Gene co-regulation is highly conserved in the evolution of eukaryotes and prokaryotes Berend Snel , * Vera van Noort , and Martijn A. Huynen Nijmegen Center for Molecular Life Sciences, University Medical Center St Radboud, p/a CMBI, Toernooiveld 1, 6525 ED Nijmegen, The Netherlands
Despite minor differences in the gene structure prediction, both gene prediction programs FGENESH and GENSCAN agree on the major features of the protein, including the presence of two WRKY domains [see Additional files 1 and 2]. Moreover, the predicted peptide sequence of the WRKY domains is identical among all the gene and domain predictions. Sequence alignment by blastn indicates that six
Mammalian Secretion of a Bacterial Protein cleaves the vector’s multiple cloning sequence upstream of the celE’ gene), and the signal peptide coding sequences, digested with the
Gene similarity networks provide tools for understanding eukaryote origins and evolution David Alvarez-Poncea,1,2, Philippe Lopezb, Eric Baptesteb, and James O. McInerneya,3,4
Gene Prediction 10/21/05 D Dobbs ISU – BCB 444/544X 1 10/21/05 D Dobbs ISU – BCB 444/544X: Gene Prediction 1 10/21/05 Gene Prediction (formerly Gene Prediction – 3)
Gene Prediction-Genotology-Lecture Slides Docsity
Ranking Gene Ontology terms for predicting non-classical
These factors explain the unique origin of eukaryotes, the absence of true evolutionary intermediates, and the evolution of sex in eukaryotes but not prokaryotes. This article was reviewed by: Eugene Koonin, William Martin, Ford Doolittle and Mark van der Giezen. For complete reports see the Reviewers’ Comments section.
eukaryotes and between eukaryotes and prokaryotes, with the present genome data these examples will not have a significant role for the prediction of functional interactions between proteins. It is possible that with the sequencing of more species such as the nematodes or yeast species in which neighboring genes do have an above-average probability of being co-regulated, the importance of gene
Superhelically Destabilized Sites in the E. coli Genome: Implications for Promoter Prediction in Prokaryotes Huiquan Wang and Craig J. Benham*
Gene Prediction 10/21/05 D Dobbs ISU – BCB 444/544X 1 10/21/05 D Dobbs ISU – BCB 444/544X: Gene Prediction 1 10/21/05 Gene Prediction (formerly Gene Prediction – 3)
Background. As more and more genomes are being sequenced, an overview of their genomic features and annotation of their functional elements, which control the expression of each gene or transcription unit of the genome, is a fundamental challenge in genomics and bioinformatics.
In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. This includes protein-coding genes as well as RNA genes, but may also include prediction of other functional elements such as regulatory regions.
Although the literature surrounding TEs in both prokaryotes and eukaryotes is clearly diverse and well developed, our understanding of this new set of mobile genetic elements, PMPs, is limited. In the sections to follow, we hope to improve this understanding in two ways. First, we substantially extend the available data regarding t he distribution of PMPs within and among sequenced prokaryotic
Despite minor differences in the gene structure prediction, both gene prediction programs FGENESH and GENSCAN agree on the major features of the protein, including the presence of two WRKY domains [see Additional files 1 and 2]. Moreover, the predicted peptide sequence of the WRKY domains is identical among all the gene and domain predictions. Sequence alignment by blastn indicates that six
This positive fashion of controlling gene expression is more common in eukaryotes than in prokaryotes. Negative control mode , where the interaction turns the genes off. In this case, a repressor protein binds the operator , a DNA sequence of approximately 20 to 25 nucleotides, which is next to the promoter or juxtaposed, and prevents the RNA polymerase from initiating transcription.
Gene prediction, especially in eukaryotic DNA since it is more complicated than prokaryotic, is an interesting problem that might be solved using machine learning. Since a DNA sequence is a chain of nucleotides, Hidden Markov Model might be used to infer gene structure. In this project I focus on predicting protein coding parts in eukaryotic DNA sequences. The problem is solved using Hidden
Gene regulation in eukaryotes. 4 questions. Practice. About this unit. You have tens of thousands of genes in your genome. Does that mean your cells express all of those genes, all the time? Not by a long shot! Even an organism as simple as a bacterium must carefully regulate gene expression, ensuring that the right genes are expressed at the right time. Learn more about the mechanisms cells
A number of examples, from eukaryotes and prokaryotes, can greatly help in improving the currently available promoter and gene prediction algorithms (40,41). METHODS Promoter sequence sets . All the promoter sequences used in this study are 1000 nt long, starting from 500 nt upstream (position −500) and extending up to 500 nt downstream (position 500) of the TSS. In order to avoid
Mammalian Secretion of a Bacterial Protein cleaves the vector’s multiple cloning sequence upstream of the celE’ gene), and the signal peptide coding sequences, digested with the
Energetics and genetics across the prokaryote-eukaryote
GeneMark.hmm eukaryotic GeneMark™ – Free gene
Eukaryotic gene prediction • Genomes are much larger than prokaryotes(10Mbp to 670 Gbp). • Low gene density. • Space between genes is very large and rich in repetitive sequences & transposable elements. • Splitting of genes by intervening noncoding sequences (introns) and joining of coding sequences (exons).
Gene similarity networks provide tools for understanding eukaryote origins and evolution David Alvarez-Poncea,1,2, Philippe Lopezb, Eric Baptesteb, and James O. McInerneya,3,4
Despite minor differences in the gene structure prediction, both gene prediction programs FGENESH and GENSCAN agree on the major features of the protein, including the presence of two WRKY domains [see Additional files 1 and 2]. Moreover, the predicted peptide sequence of the WRKY domains is identical among all the gene and domain predictions. Sequence alignment by blastn indicates that six
Gene prediction in eukaryotes is notoriously difficult; it is not even a straightforward business to decide how to count and, in humans at least, there is considerable discrepancy between different gene prediction methods . Clearly, phenomena such as alternative splicing and post-translational modifications of genes must also be taken into account. Accurate prediction of alternative gene
System I was apparently ancestral to all eukaryotes and was functionally replaced by the HCCS (which is absent in prokaryotes), but whether this happened once or multiple times is an unresolved debate, the implications of which have been hypothesized to apply to rooting the tree of eukaryotes
A number of examples, from eukaryotes and prokaryotes, can greatly help in improving the currently available promoter and gene prediction algorithms (40,41). METHODS. Promoter sequence sets . All the promoter sequences used in this study are 1000 nt long, starting from 500 nt upstream (position −500) and extending up to 500 nt downstream (position 500) of the TSS. In order to avoid
(formerly Gene Prediction – 1) • Eukaryotes vs prokaryotes • Cells • Genome organization • Gene structure 10/19/05 D Dobbs ISU – BCB 444/544X: Gene Regulation 6 Eukaryotes vs Prokaryotes Eukaryotic cells are characterized by membrane-bound compartments, which are absent in prokaryotes. “Typical” human & bacterial cells drawn to scale. BIOS c i en tf Publ shrLd,1 9 Brown …
Although gene prediction has been the subject of several excellent review papers (Do and Choi, 2006, Brent, 2007, Flicek, 2007); the current study is designed to appeal to both the expert and non-expert reader alike; providing a concise overview of the current status of eukaryote gene prediction models, beginning with a brief overview of sensor recognition and gene-finders, their strengths and
Introduction Gene Prediction. Gene prediction is the process of identifying specific regions of genomic DNA that encode for genes. A gene is the entire DNA sequence necessary for production of a functional polypeptide or RNA.
Gene Prediction by Homology •In both prokaryotes and eukaryotes, genes can occur in any reading frame and on either strand (always in the 5′ to 3′ direction) •Also, not all genes are coding. Annotation of a segment of the E. coligenome. Observations •Non-coding genes such as tRNAsdo not have corresponding proteins •These have conserved structures that aid in their identification
Gene prediction, especially in eukaryotic DNA since it is more complicated than prokaryotic, is an interesting problem that might be solved using machine learning. Since a DNA sequence is a chain of nucleotides, Hidden Markov Model might be used to infer gene structure. In this project I focus on predicting protein coding parts in eukaryotic DNA sequences. The problem is solved using Hidden
Computational Model for Prokaryotic and Eukaryotic Gene Prediction 345 There is one-to-one correspondence between the nucleotides used to make ribonu- cleic acid (G, A, U and C where ―U is an abbreviation for Uracil) and the nucleotide
Mammalian Secretion of a Bacterial Protein cleaves the vector’s multiple cloning sequence upstream of the celE’ gene), and the signal peptide coding sequences, digested with the
Evaluation of gene prediction methods for prokaryotes Master’s thesis Stian Engebretsen 15th August 2012. Abstract There are currently over 2,000 completed prokaryotic sequences available. In these DNA sequences the genes lay hidden. The genes codes for proteins that forms the foundation for all organisms. Several programs that try to locate these genes have been developed. However, little
Biodata of Mario Stanke author of “Computational Gene
UniLoc A universal protein localization site predictor
a presentation regarding gene prediction Download as PPTX, PDF, TXT or read online from Scribd
Although the literature surrounding TEs in both prokaryotes and eukaryotes is clearly diverse and well developed, our understanding of this new set of mobile genetic elements, PMPs, is limited. In the sections to follow, we hope to improve this understanding in two ways. First, we substantially extend the available data regarding t he distribution of PMPs within and among sequenced prokaryotic
Algorithms in Bioinformatics II, SS’07, ZBIT, C. Dieterich, April 17, 2007 193 12 Gene Prediction This chapter is based on the following sources, which are all recommended reading:
System I was apparently ancestral to all eukaryotes and was functionally replaced by the HCCS (which is absent in prokaryotes), but whether this happened once or multiple times is an unresolved debate, the implications of which have been hypothesized to apply to rooting the tree of eukaryotes
In eukaryotes, a gene is a combination of coding segments (exons) that are interrupted by non-coding segments (introns) This makes computational gene prediction in eukaryotes even more
Even if the gene model is fixed, the sources of information taken into account be EUGÈNE for prediction are extremely varied and can be easily extended by creating so-called plugins. Currently, E …
Gene similarity networks provide tools for understanding eukaryote origins and evolution David Alvarez-Poncea,1,2, Philippe Lopezb, Eric Baptesteb, and James O. McInerneya,3,4
A simple gene structure Although introns do exist in prokaryotes, they are extremely rare and often ignored by gene prediction tools. The relative simplicity of bacterial gene
divided into two, namely, Gene Prediction in Prokaryotes and in Eukaryotes. 5 Gene Prediction in Prokaryotes 5.1 Understanding prokaryotic gene structure The knowledge of gene structure is very important when we set out to solve the problem of gene prediction. The gene structure of Prokaryotes can be captured in terms of the following characteristics Promoter Elements The process of gene
(formerly Gene Prediction – 1) • Eukaryotes vs prokaryotes • Cells • Genome organization • Gene structure 10/19/05 D Dobbs ISU – BCB 444/544X: Gene Regulation 6 Eukaryotes vs Prokaryotes Eukaryotic cells are characterized by membrane-bound compartments, which are absent in prokaryotes. “Typical” human & bacterial cells drawn to scale. BIOS c i en tf Publ shrLd,1 9 Brown …
Automatic gene prediction is one of the essential issues in bioinformatics. Many approaches have been proposed and a lot of tools have been developed. This paper compiles information about some of
This positive fashion of controlling gene expression is more common in eukaryotes than in prokaryotes. Negative control mode , where the interaction turns the genes off. In this case, a repressor protein binds the operator , a DNA sequence of approximately 20 to 25 nucleotides, which is next to the promoter or juxtaposed, and prevents the RNA polymerase from initiating transcription.
Gene regulation in eukaryotes. 4 questions. Practice. About this unit. You have tens of thousands of genes in your genome. Does that mean your cells express all of those genes, all the time? Not by a long shot! Even an organism as simple as a bacterium must carefully regulate gene expression, ensuring that the right genes are expressed at the right time. Learn more about the mechanisms cells
and evaluated with non-redundant proteins of eukaryotes and prokaryotes, whereas KnowPred site was trained and evaluated with a set of redundaneukaryotic t proteins.
Gene Prediction Group Computational Genomics 2016
Gene Prediction in Eukaryotes Using Machine Learning
Gnomon – NCBI eukaryotic gene prediction tool A. Souvorov ∗, Y. Kapustin, B. Kiryutin, V. Chetvernin, T. Tatusova, and D. Lipman National Center for Biotechnology Information, Bethesda MD February 25, 2010 ∗To whom correspondence should be addressed. 1 . 1 Methods NCBI gene prediction is a combination of homology searching with ab initio modeling. The use of ab initio is threefold: a) we
Gene prediction, especially in eukaryotic DNA since it is more complicated than prokaryotic, is an interesting problem that might be solved using machine learning. Since a DNA sequence is a chain of nucleotides, Hidden Markov Model might be used to infer gene structure. In this project I focus on predicting protein coding parts in eukaryotic DNA sequences. The problem is solved using Hidden
Although the literature surrounding TEs in both prokaryotes and eukaryotes is clearly diverse and well developed, our understanding of this new set of mobile genetic elements, PMPs, is limited. In the sections to follow, we hope to improve this understanding in two ways. First, we substantially extend the available data regarding t he distribution of PMPs within and among sequenced prokaryotic
Contains fulltext : 30111_compgeofe.pdf (publisher’s version ) (Open Access)This thesis focuses on developing comparative genomics methods in eukaryotes, with an emphasis on applications for gene function prediction and regulatory element detection. In the past, methods have been developed to
Gene Prediction by Homology •In both prokaryotes and eukaryotes, genes can occur in any reading frame and on either strand (always in the 5′ to 3′ direction) •Also, not all genes are coding. Annotation of a segment of the E. coligenome. Observations •Non-coding genes such as tRNAsdo not have corresponding proteins •These have conserved structures that aid in their identification
Mammalian Secretion of a Bacterial Protein cleaves the vector’s multiple cloning sequence upstream of the celE’ gene), and the signal peptide coding sequences, digested with the
A number of examples, from eukaryotes and prokaryotes, can greatly help in improving the currently available promoter and gene prediction algorithms (40,41). METHODS Promoter sequence sets . All the promoter sequences used in this study are 1000 nt long, starting from 500 nt upstream (position −500) and extending up to 500 nt downstream (position 500) of the TSS. In order to avoid
The major differences between gene expression of prokaryotes and eukaryotes are that prokaryotes have single circular chromosome, whereas eukaryotes have many bar shaped chromosomes. The gene expression for eukaryotes completes in two different steps, one with transcription than with translation. In eukaryotes the open reading frames are longer as compared to open reading frames of prokaryotes
Birth Death and Diversi cation of Mobile Promoters in
An overview of the current status of eukaryote gene
Gene prediction, especially in eukaryotic DNA since it is more complicated than prokaryotic, is an interesting problem that might be solved using machine learning. Since a DNA sequence is a chain of nucleotides, Hidden Markov Model might be used to infer gene structure. In this project I focus on predicting protein coding parts in eukaryotic DNA sequences. The problem is solved using Hidden
Findings. We present a software package and a web server for predicting the subcellular localization of protein sequences based on the ngLOC method. ngLOC is an n-gram-based Bayesian classifier that predicts subcellular localization of proteins both in prokaryotes and eukaryotes.
RNA Polymerase: Prokaryotes have one RNA polymerase that handles all transcription while eukaryotes have 3. The prokaryotic RNA polymerase is much …
Gene Prediction 10/21/05 D Dobbs ISU – BCB 444/544X 1 10/21/05 D Dobbs ISU – BCB 444/544X: Gene Prediction 1 10/21/05 Gene Prediction (formerly Gene Prediction – 3)
Algorithms in Bioinformatics II, SS’07, ZBIT, C. Dieterich, April 17, 2007 193 12 Gene Prediction This chapter is based on the following sources, which are all recommended reading:
Gene Prediction Ppt – Download as Powerpoint Presentation (.ppt), PDF File (.pdf), Text File (.txt) or view presentation slides online. Scribd is the world’s largest social reading and publishing site.
In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. This includes protein-coding genes as well as RNA genes, but may also include prediction of other functional elements such as regulatory regions.
also reported in a few eukaryotes [2, 12]. The genes transcribed in an operon are functionally related The genes transcribed in an operon are functionally related and the proteins are part of the same protein complex or metabolic pathway [14].
Background. As more and more genomes are being sequenced, an overview of their genomic features and annotation of their functional elements, which control the expression of each gene or transcription unit of the genome, is a fundamental challenge in genomics and bioinformatics.
and evaluated with non-redundant proteins of eukaryotes and prokaryotes, whereas KnowPred site was trained and evaluated with a set of redundaneukaryotic t proteins.
Two Approaches to Gene Prediction An Introduction to Bioinformatics Algorithms www.bioalgorithms.info If you could compare the day’s news in English, side-by-side
(PDF) Computational Methods for Gene Finding in Prokaryotes
Genome and protein evolution in eukaryotes ScienceDirect
Algorithms in Bioinformatics II, SS’07, ZBIT, C. Dieterich, April 17, 2007 193 12 Gene Prediction This chapter is based on the following sources, which are all recommended reading:
and evaluated with non-redundant proteins of eukaryotes and prokaryotes, whereas KnowPred site was trained and evaluated with a set of redundaneukaryotic t proteins.
This positive fashion of controlling gene expression is more common in eukaryotes than in prokaryotes. Negative control mode , where the interaction turns the genes off. In this case, a repressor protein binds the operator , a DNA sequence of approximately 20 to 25 nucleotides, which is next to the promoter or juxtaposed, and prevents the RNA polymerase from initiating transcription.
A simple gene structure Although introns do exist in prokaryotes, they are extremely rare and often ignored by gene prediction tools. The relative simplicity of bacterial gene
also reported in a few eukaryotes [2, 12]. The genes transcribed in an operon are functionally related The genes transcribed in an operon are functionally related and the proteins are part of the same protein complex or metabolic pathway [14].
unicellular eukaryotes (1, 2). Also, with the exception of deterio- rating genomes of some parasitic bacteria, the prokaryotic genomes are highly compact, with densely packed protein-coding genes and a low fraction of noncoding sequences (3). The small genome size is thought to be selected for fast re plication, whereas the high gene density additionally facilitates coregulation of gene
These factors explain the unique origin of eukaryotes, the absence of true evolutionary intermediates, and the evolution of sex in eukaryotes but not prokaryotes. This article was reviewed by: Eugene Koonin, William Martin, Ford Doolittle and Mark van der Giezen. For complete reports see the Reviewers’ Comments section.
Anecdotal evidence shows promoters being reused separate from their downstream gene, thus providing a mechanism for the efficient and rapid rewiring of a gene’s transcriptional regulation. We have identified over 4000 groups of highly similar promoters using a conservative sequence similarity search in all fully sequenced prokaryotic genomes.
Despite minor differences in the gene structure prediction, both gene prediction programs FGENESH and GENSCAN agree on the major features of the protein, including the presence of two WRKY domains [see Additional files 1 and 2]. Moreover, the predicted peptide sequence of the WRKY domains is identical among all the gene and domain predictions. Sequence alignment by blastn indicates that six
Eukaryotic gene prediction • Genomes are much larger than prokaryotes(10Mbp to 670 Gbp). • Low gene density. • Space between genes is very large and rich in repetitive sequences & transposable elements. • Splitting of genes by intervening noncoding sequences (introns) and joining of coding sequences (exons).
Contains fulltext : 30111_compgeofe.pdf (publisher’s version ) (Open Access)This thesis focuses on developing comparative genomics methods in eukaryotes, with an emphasis on applications for gene function prediction and regulatory element detection. In the past, methods have been developed to
(formerly Gene Prediction – 1) • Eukaryotes vs prokaryotes • Cells • Genome organization • Gene structure 10/19/05 D Dobbs ISU – BCB 444/544X: Gene Regulation 6 Eukaryotes vs Prokaryotes Eukaryotic cells are characterized by membrane-bound compartments, which are absent in prokaryotes. “Typical” human & bacterial cells drawn to scale. BIOS c i en tf Publ shrLd,1 9 Brown …
In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. This includes protein-coding genes as well as RNA genes, but may also include prediction of other functional elements such as regulatory regions.
GeneMark.hmm eukaryotic Eukaryotic GeneMark.hmm with supervised training was not described in any publication as a stand alone algorithm. However, it was used and evaluated in several projects e.g. in Pavy et al. “Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thaliana sequences” Bioinformatics 1999, 15, 887-99.
PromBase a web resource for various genomic features and
Comparative Analysis of Gene Prediction Tools RAST
Despite minor differences in the gene structure prediction, both gene prediction programs FGENESH and GENSCAN agree on the major features of the protein, including the presence of two WRKY domains [see Additional files 1 and 2]. Moreover, the predicted peptide sequence of the WRKY domains is identical among all the gene and domain predictions. Sequence alignment by blastn indicates that six
Gene prediction in eukaryotes is notoriously difficult; it is not even a straightforward business to decide how to count and, in humans at least, there is considerable discrepancy between different gene prediction methods . Clearly, phenomena such as alternative splicing and post-translational modifications of genes must also be taken into account. Accurate prediction of alternative gene
Homology-based gene prediction Similarity Searches (e.g. BLAST, BLAT) Genome Browsers RNA evidence (ESTs) Ab initio gene prediction Gene prediction programs Prokaryotes ORF identification Eukaryotes
eukaryotes and between eukaryotes and prokaryotes, with the present genome data these examples will not have a significant role for the prediction of functional interactions between proteins. It is possible that with the sequencing of more species such as the nematodes or yeast species in which neighboring genes do have an above-average probability of being co-regulated, the importance of gene
Prokaryotic vs Eukaryotic Gene Prediction ⚫Simpler in Prokaryotes than in Eukaryotes: ⚫Prokaryotic genomes are gene-rich with their genomes comprised with at least 90% coding
Two Approaches to Gene Prediction An Introduction to Bioinformatics Algorithms www.bioalgorithms.info If you could compare the day’s news in English, side-by-side
GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses John Besemer1 and Mark Borodovsky1,2
Superhelically Destabilized Sites in the E. coli Genome: Implications for Promoter Prediction in Prokaryotes Huiquan Wang and Craig J. Benham*
10/19/05 Gene Prediction & Regulation Gene ERegulationu
Comparative Genomics of Eukaryotes. CORE
a presentation regarding gene prediction Download as PPTX, PDF, TXT or read online from Scribd
also reported in a few eukaryotes [2, 12]. The genes transcribed in an operon are functionally related The genes transcribed in an operon are functionally related and the proteins are part of the same protein complex or metabolic pathway [14].
A promoter is a regulatory region of DNA located upstream (towards the 5′ region) of of a gene, providing a control point for regulated gene transcription. The promoter contains specific DNA sequences that are recognized by proteins known as transcription factors.
eukaryotes and between eukaryotes and prokaryotes, with the present genome data these examples will not have a significant role for the prediction of functional interactions between proteins. It is possible that with the sequencing of more species such as the nematodes or yeast species in which neighboring genes do have an above-average probability of being co-regulated, the importance of gene
System I was apparently ancestral to all eukaryotes and was functionally replaced by the HCCS (which is absent in prokaryotes), but whether this happened once or multiple times is an unresolved debate, the implications of which have been hypothesized to apply to rooting the tree of eukaryotes
Mammalian Secretion of a Bacterial Protein cleaves the vector’s multiple cloning sequence upstream of the celE’ gene), and the signal peptide coding sequences, digested with the
Eukaryotic gene prediction • Genomes are much larger than prokaryotes(10Mbp to 670 Gbp). • Low gene density. • Space between genes is very large and rich in repetitive sequences & transposable elements. • Splitting of genes by intervening noncoding sequences (introns) and joining of coding sequences (exons).
GeneMark.hmm eukaryotic Eukaryotic GeneMark.hmm with supervised training was not described in any publication as a stand alone algorithm. However, it was used and evaluated in several projects e.g. in Pavy et al. “Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thaliana sequences” Bioinformatics 1999, 15, 887-99.
In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. This includes protein-coding genes as well as RNA genes, but may also include prediction of other functional elements such as regulatory regions.
GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses John Besemer1 and Mark Borodovsky1,2
A New Lineage of Eukaryotes Illuminates Early
Homology-based gene prediction Similarity Searches (e.g. BLAST, BLAT) Genome Browsers RNA evidence (ESTs) Ab initio gene prediction Gene prediction programs Prokaryotes ORF identification Eukaryotes
Survey and research proposal on Computational methods for
Superhelically Destabilized Sites in the E. coli Genome
Read “Ranking Gene Ontology terms for predicting non-classical secretory proteins in eukaryotes and prokaryotes, Journal of Theoretical Biology” on DeepDyve, the largest online rental service for scholarly research with thousands of academic publications available at your fingertips.
CHAPTER 9 GENE PREDICTION gdancik.github.io
Conservation of gene co-regulation in prokaryotes and eukaryotes Sarah A. Teichmann and M. Madan Babu Genes that are part of the same operon in prokaryotes,or have the same expression pattern in eukaryotes,are transcriptionally co-regulated.If genes are consistently co-regulated across distantly related organisms,the genes have closely associated functions.It has been shown …
Promoter reuse in prokaryotes Mobile Genetic Elements
GeneMark web software for gene finding in prokaryotes
Survey and research proposal on Computational methods for gene prediction in Eukaryotes – A Report Dhruv Jain 1. ABSTRACT The rising popularity of genome sequencing in …
12 Gene Prediction Algorithms in Bioinformatics
Background. As more and more genomes are being sequenced, an overview of their genomic features and annotation of their functional elements, which control the expression of each gene or transcription unit of the genome, is a fundamental challenge in genomics and bioinformatics.
Research News Conservation of gene co-regulation in
Homology-based gene prediction Similarity Searches (e.g. BLAST, BLAT) Genome Browsers RNA evidence (ESTs) Ab initio gene prediction Gene prediction programs Prokaryotes ORF identification Eukaryotes
Gene Prediction in Eukaryotes Using Machine Learning
Comparative Analysis of Gene Prediction Tools RAST
A natural barrier to lateral gene transfer from
Automatic gene prediction is one of the essential issues in bioinformatics. Many approaches have been proposed and a lot of tools have been developed. This paper compiles information about some of
Biodata of Mario Stanke author of “Computational Gene
Gene Prediction (1) Promoter (Genetics) Gene
Ranking Gene Ontology terms for predicting non-classical
Gnomon – NCBI eukaryotic gene prediction tool A. Souvorov ∗, Y. Kapustin, B. Kiryutin, V. Chetvernin, T. Tatusova, and D. Lipman National Center for Biotechnology Information, Bethesda MD February 25, 2010 ∗To whom correspondence should be addressed. 1 . 1 Methods NCBI gene prediction is a combination of homology searching with ab initio modeling. The use of ab initio is threefold: a) we
EUGÈNE an open gene finder for eukaryotes and prokaryotes
Genome Characteristics and Annotation Rice University
Although gene prediction has been the subject of several excellent review papers (Do and Choi, 2006, Brent, 2007, Flicek, 2007); the current study is designed to appeal to both the expert and non-expert reader alike; providing a concise overview of the current status of eukaryote gene prediction models, beginning with a brief overview of sensor recognition and gene-finders, their strengths and
Energetics and genetics across the prokaryote-eukaryote
Promoter region in Eukaryotes and Prokaryotes
Promoter reuse in prokaryotes Mobile Genetic Elements
Gene Prediction Ppt – Download as Powerpoint Presentation (.ppt), PDF File (.pdf), Text File (.txt) or view presentation slides online. Scribd is the world’s largest social reading and publishing site.
COMPUTATIONAL GENE PREDICTION
Introduction Gene Prediction. Gene prediction is the process of identifying specific regions of genomic DNA that encode for genes. A gene is the entire DNA sequence necessary for production of a functional polypeptide or RNA.
Genome and protein evolution in eukaryotes ScienceDirect
The major differences between gene expression of prokaryotes and eukaryotes are that prokaryotes have single circular chromosome, whereas eukaryotes have many bar shaped chromosomes. The gene expression for eukaryotes completes in two different steps, one with transcription than with translation. In eukaryotes the open reading frames are longer as compared to open reading frames of prokaryotes
COMPUTATIONAL GENE PREDICTION
Gene Prediction Ppt Gene Messenger Rna
Genome Analysis Inversions and the dynamics of eukaryotic
RNA Polymerase: Prokaryotes have one RNA polymerase that handles all transcription while eukaryotes have 3. The prokaryotic RNA polymerase is much …
Superhelically Destabilized Sites in the E. coli Genome
UniLoc A universal protein localization site predictor
CHAPTER 9 GENE PREDICTION gdancik.github.io
These factors explain the unique origin of eukaryotes, the absence of true evolutionary intermediates, and the evolution of sex in eukaryotes but not prokaryotes. This article was reviewed by: Eugene Koonin, William Martin, Ford Doolittle and Mark van der Giezen. For complete reports see the Reviewers’ Comments section.
EUGÈNE an open gene finder for eukaryotes and prokaryotes
UniLoc A universal protein localization site predictor
Clustering Algorithm Approach for Prokaryotic and
Hot, Toxic Eukaryote. Unusually, the single-celled eukaryote red alga, Galdieria sulphuraria, can thrive in hot, acidic springs. This organism is endowed with extraordinary metabolic talents and can consume a variety of strange carbohydrates, as well as turn on photosynthesis when the food runs out.
GeneMark.hmm eukaryotic GeneMark™ – Free gene
ngLOC software and web server for predicting protein
System I was apparently ancestral to all eukaryotes and was functionally replaced by the HCCS (which is absent in prokaryotes), but whether this happened once or multiple times is an unresolved debate, the implications of which have been hypothesized to apply to rooting the tree of eukaryotes
Predictionof*Prokaryotic*andEukaryotic*Promoters*Using
COMPUTATIONAL GENE PREDICTION
Comparative Analysis of Gene Prediction Tools RAST
The Significance of Intron Distributions for Gene Prediction In contrast to prokaryotes, in which each gene is encoded by a contiguous seg-ment of DNA, most eukaryotic genomes contain introns, which interrupt the genes. However, there also exist a few eukaryotes in which most genes are intron-less such as the red alga Cyanidioschyzon merolae (Nozaki et al., 2007) and the yeast Saccharomyces
A New Lineage of Eukaryotes Illuminates Early
(PDF) Computational Methods for Gene Finding in Prokaryotes
Prokaryotic vs Eukaryotic Gene Prediction ⚫Simpler in Prokaryotes than in Eukaryotes: ⚫Prokaryotic genomes are gene-rich with their genomes comprised with at least 90% coding
Gene Prediction Background & Strategy Compgenomics2017
System I was apparently ancestral to all eukaryotes and was functionally replaced by the HCCS (which is absent in prokaryotes), but whether this happened once or multiple times is an unresolved debate, the implications of which have been hypothesized to apply to rooting the tree of eukaryotes
Promoter region in Eukaryotes and Prokaryotes
This comprises cellular logistics in prokaryotes as well as eukaryotes, and it is this aspect of cytokinesis which is the subject of this review. This review consists of three parts. First, I will present a brief overview of cytokinesis on the cellular and subcellular levels.
Gene Prediction (1) Promoter (Genetics) Gene
GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses John Besemer1 and Mark Borodovsky1,2
Comparative Analysis of Gene Prediction Tools RAST
GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses John Besemer1 and Mark Borodovsky1,2
A natural barrier to lateral gene transfer from
GeneMark gene prediction
Gene co-regulation is highly conserved in the evolution of
Eukaryotic gene prediction • Genomes are much larger than prokaryotes(10Mbp to 670 Gbp). • Low gene density. • Space between genes is very large and rich in repetitive sequences & transposable elements. • Splitting of genes by intervening noncoding sequences (introns) and joining of coding sequences (exons).
Gene co-regulation is highly conserved in the evolution of
Gene similarity networks provide tools for understanding
Comparative Analysis of Gene Prediction Tools RAST
We present a WWW server for AUGUSTUS, a software for gene prediction in eukaryotic genomic sequences that is based on a generalized hidden Markov model, a probabilistic model of a sequence and its gene structure. The web server allows the user to impose constraints on the predicted gene …
Prokaryotic Gene Prediction Using GeneMark and GeneMark
Conservation of gene co-regulation in prokaryotes and eukaryotes Sarah A. Teichmann and M. Madan Babu Genes that are part of the same operon in prokaryotes,or have the same expression pattern in eukaryotes,are transcriptionally co-regulated.If genes are consistently co-regulated across distantly related organisms,the genes have closely associated functions.It has been shown …
Energetics and genetics across the prokaryote-eukaryote
The WRKY transcription factor superfamily its origin in
gene structure prediction and annotation is the fundamental step towards the understanding of genome function. A A large number of gene prediction tool and pipeline have …
Superhelically Destabilized Sites in the E. coli Genome
Gene Prediction in Eukaryotes Using Machine Learning
CCIS 169 Computational Model for Prokaryotic and
A number of examples, from eukaryotes and prokaryotes, can greatly help in improving the currently available promoter and gene prediction algorithms (40,41). METHODS Promoter sequence sets . All the promoter sequences used in this study are 1000 nt long, starting from 500 nt upstream (position −500) and extending up to 500 nt downstream (position +500) of the TSS. In order to avoid
ngLOC software and web server for predicting protein
Clustering Algorithm Approach for Prokaryotic and
CCIS 169 Computational Model for Prokaryotic and
These factors explain the unique origin of eukaryotes, the absence of true evolutionary intermediates, and the evolution of sex in eukaryotes but not prokaryotes. This article was reviewed by: Eugene Koonin, William Martin, Ford Doolittle and Mark van der Giezen. For complete reports see the Reviewers’ Comments section.
Research News Conservation of gene co-regulation in
Gnomon NCBI eukaryotic gene prediction tool
This positive fashion of controlling gene expression is more common in eukaryotes than in prokaryotes. Negative control mode , where the interaction turns the genes off. In this case, a repressor protein binds the operator , a DNA sequence of approximately 20 to 25 nucleotides, which is next to the promoter or juxtaposed, and prevents the RNA polymerase from initiating transcription.
(PDF) Computational Methods for Gene Finding in Prokaryotes
Prokaryotic promoters BiOS
In this study, we investigated the possible origin of β-CA gene sequences in protozoans, insects, and nematodes by HGT from ancestral prokaryotes using phylogenetics, prediction of subcellular localization, and identification of β-CA, transposase, integrase, and resolvase genes on the MGEs of bacteria. We also structurally analyzed β-CAs from protozoans, insects, and nematodes and their
CHAPTER 9 GENE PREDICTION gdancik.github.io
Gene Prediction Background & Strategy Compgenomics2017
Gene Prediction Statistical Approaches UCSD CSE
Read “Ranking Gene Ontology terms for predicting non-classical secretory proteins in eukaryotes and prokaryotes, Journal of Theoretical Biology” on DeepDyve, the largest online rental service for scholarly research with thousands of academic publications available at your fingertips.
Energetics and genetics across the prokaryote-eukaryote
Clustering Algorithm Approach for Prokaryotic and